Actinobacillus succinogenes genetically engineered bacterium and construction method and application thereof
A technology of genetically engineered bacteria and actinomycetes, applied in the field of metabolic engineering, can solve the problems of increased cost, easy death, weak acid resistance of Actinobacillus succinates, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
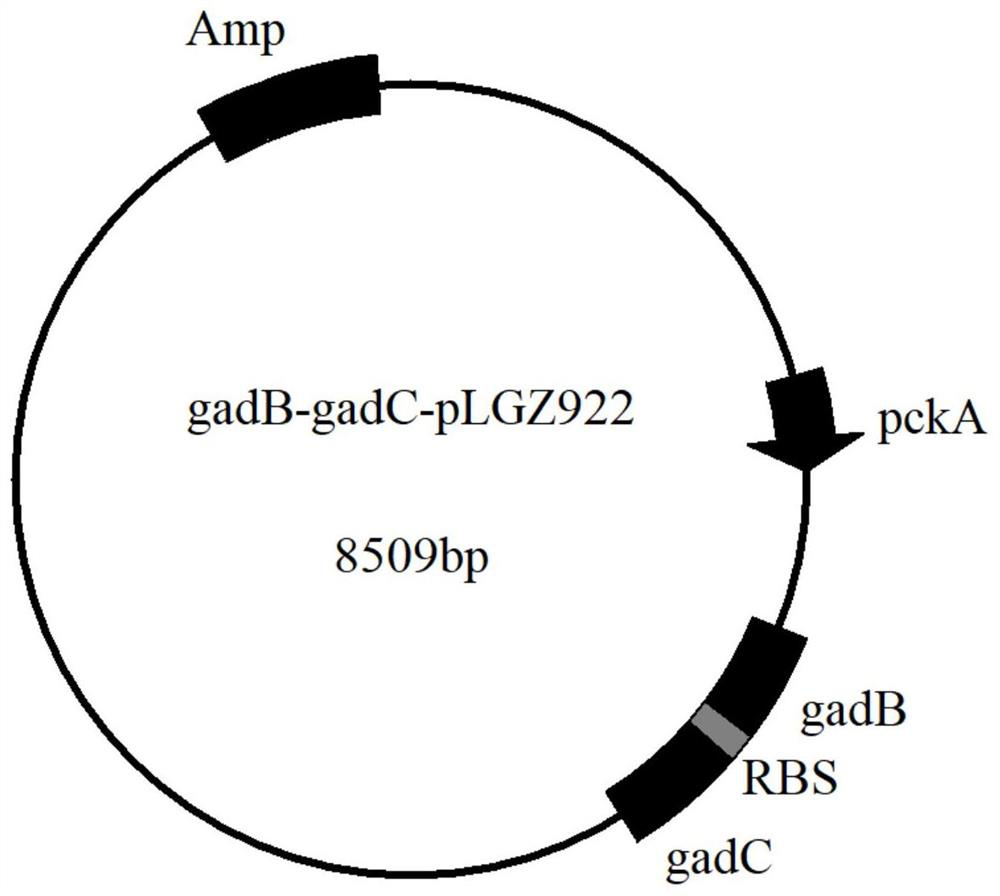
[0026] Example 1: Overexpression strains gadB-WS058, gadB-gadC-WS058 and gadB △C11 - build of gadC-WS058
[0027] This embodiment specifically illustrates the overexpression strains gadB-WS058, gadB-gadC-WS058 and gadB △C11 The construction process of -gadC-WS058, the specific steps are as follows:
[0028] Use the Ezvp Column Bacterial Genomic DNA Extraction Kit (Shanghai Shenggong) to extract the genome of Lactobacillus brinneri preserved in the laboratory, and use it as a template to use gadB gene primers (F1 and F2) / gadB △C11 (F1 and F5) and gadC gene primers (F3 and F4) PCR amplified gadB and gadC / gadB △C11 Fragment where gadB / gadB △C11 and gadC gene fragments are about 1449bp and 1521bp respectively, and then use primers F1 and F4 to amplify gadB-gadC / gadB by fusion PCR △C11 - The gadC fragment, whose size is about 2979bp, obtains the Gad system gene expression cassette.
[0029] Primer sequence (SEQ ID NO.7~11):
[0030] F1: TTATCAATGAGGTGA TCTAGA ATGAGTGAAAAAAA...
Embodiment 2
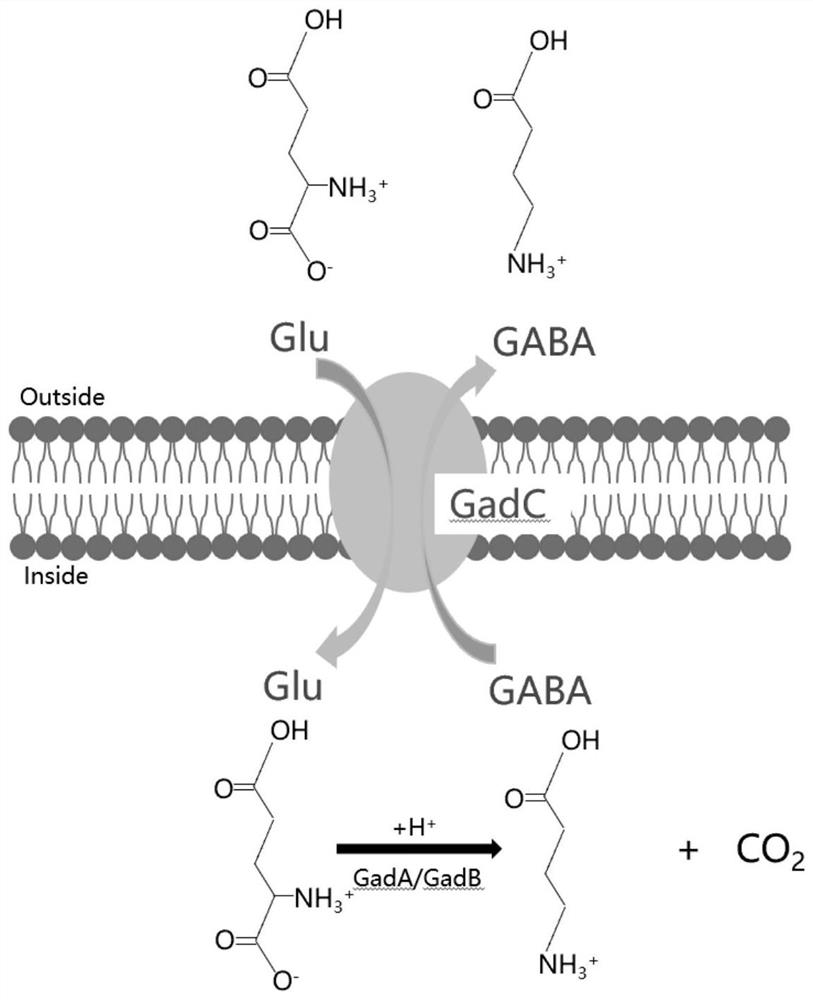
[0036] Embodiment 2: Enzyme activity assay of Gad system in Actinobacillus succinate
[0037] This embodiment specifically illustrates whether there is enzyme activity after the Gad system is introduced into Actinobacillus succinogenes, and the specific determination method is as follows:
[0038] Take out the strains WS058, pLGZ922-WS058, gadB preserved in glycerol tubes from the refrigerator △C11 -gadC-WS058, gadB-gadC-WS058, melted at room temperature (take 750 μL) into TSB liquid (50ml / 100ml) medium, anaerobic culture at 37°C for 30h.
[0039] Centrifuge the cultured bacterial solution at 6000 rpm at 4°C for 10 minutes, collect the bacterial cells, wash with water once, add 10 mL of distilled water, suspend the bacterial cells, and obtain a sample solution.
[0040] 0.2mol·L -1 Sodium acetate-acetic acid buffer (pH 4.6) was used as the reaction buffer, and 60 mM sodium glutamate and 0.6 mM pyridoxal phosphate were added to prepare the substrate buffer. Mix 1 mL of the sub...
Embodiment 3
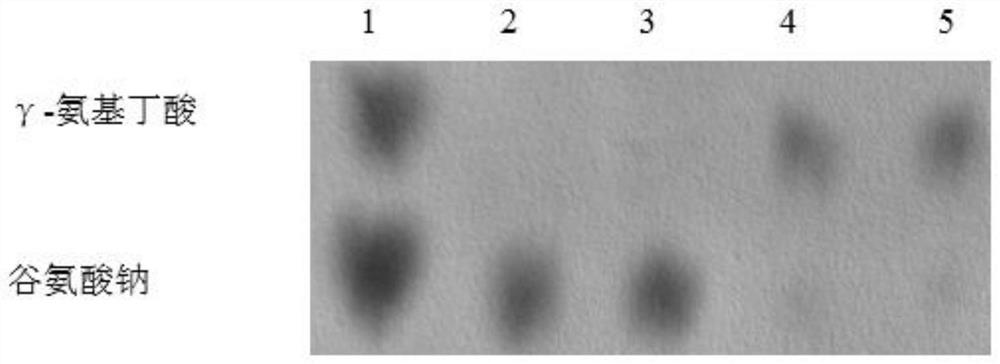
[0043] Embodiment 3: the mensuration of acid resistance
[0044] The present embodiment specifically illustrates the influence of Actinobacillus succinate into the Gad system on the acid resistance of the bacterial strain, and the specific assay method is as follows:
[0045] Take out the strains stored in glycerol tubes from the refrigerator, melt them at room temperature, and activate them by streaking. Pick a single colony and place them in TSB medium for anaerobic culture at 37°C for 24 hours; then absorb 30 mL of bacterial liquid and add them to 50 mL centrifuge tubes, centrifuge at 8000 rpm After 10 minutes, the supernatant was discarded, and 30 mL of sterile water was added to suspend the bacteria, and then 5 mL were drawn and added to TSB medium containing 45 mL of pH 4.6 with different concentrations of sodium glutamate, and cultured anaerobically at 37 °C for 1.5 h. Draw 1 mL, wash it with water once, and dilute it according to a certain ratio, then draw 5 μL and spo...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com