Preparation method of multi-fluorescent nucleic acid probe and application thereof
A fluorescent nucleic acid and probe technology, which is applied in the field of preparation of multi-fluorescent nucleic acid probes, can solve the problems of limiting the sensitivity of probes and affecting the detection limit of biosensors, and achieves enhanced sensitivity, short time required, and simple preparation methods Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0021] Embodiment 1 prepares the multi-fluorescence nucleic acid probe method for detecting EV-71 virus
[0022] A. Preparation of double-stranded DNA:
[0023] Template sequences, upstream primers, and downstream primers were all synthesized by Takara Biological Company.
[0024] Template sequence: 5'-GAG CAG TCA CAG TCC AGA AGG GCA TGT CAG GGC TTG GAT ACCTCG CAT TCA CCC TTG CAC GAT AC-3';
[0025] Upstream primer: 5'-GAG CAG TCA CAG TCC AGA AG-3' (phosphorylated at the 5' end)
[0026] Downstream primer: 5’-GTA TCG TGC AAG GGT GAA TGC-3’
[0027] PCR reaction system
[0028]
[0029]
[0030] The PCR reaction was carried out according to the above PCR reaction system, the amplification conditions were: 95°C, 30s; 55°C, 30s; 72°C, 30s, after 40 cycles, 72°C, 5min. The amplified product is purified by the kit to obtain pure double-stranded DNA.
[0031] B. Preparation of probe single strand:
[0032] The double-stranded DNA obtained in step A was mixed with Lambda ...
Embodiment 2
[0037] Comparison of the double-stranded DNA synthesized by embodiment 2EdUTP and dTTP
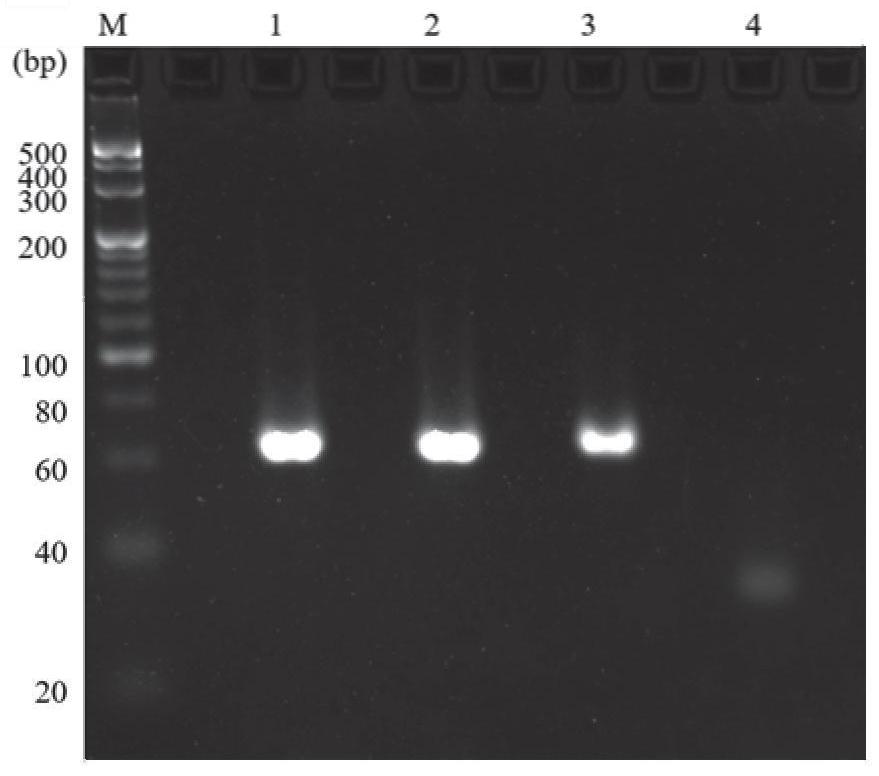
[0038] According to step A in Example 1, replace EdUTP with dTTP to obtain double-stranded DNA. The double-stranded DNA, the high-density alkyne double-stranded DNA synthesized by EdUTP obtained in Example 1, the purified high-density alkyne double-stranded DNA, and the probe chain were analyzed by agarose gel electrophoresis. Proceed as follows:
[0039] Gel production: Add accurately weighed agarose powder to a conical flask with 60mL of 1X TBE buffer, heat and dissolve in a microwave oven, cool to about 50-60°C, add ethidium bromide, and mix well Introduce into the glue mold and let the glue solidify at room temperature.
[0040] Sample loading: Take an appropriate amount of sample and mix with 6× sample buffer, and the sample volume for each sample tank is 10 μL.
[0041] Electrophoresis: After adding the sample, close the cover of the electrophoresis tank and turn on the power imme...
Embodiment 3
[0043] The comparison of embodiment 3 synthetic probe strands and standard probes
[0044] The purchased standard probe sequence is: 5'-GTA TCG TGC AAG GGT GAA TGC GAG GTA TCC AAGCCC TGA CAT GCC CTT CTG GAC TGT GAC TGC TC-3', a fluorescent molecule is attached to the 5' end;
[0045] The sequence of the multi-fluorescent nucleic acid probe synthesized by the present invention is: 5'-GTA TCG TGC AAG GGT GAA TGC GAGGTA TCC AAG CCC TGA CAT GCC CTT CTG GAC TGT GAC TGC TC-3', and there are multiple fluorescent molecules attached to the DNA chain;
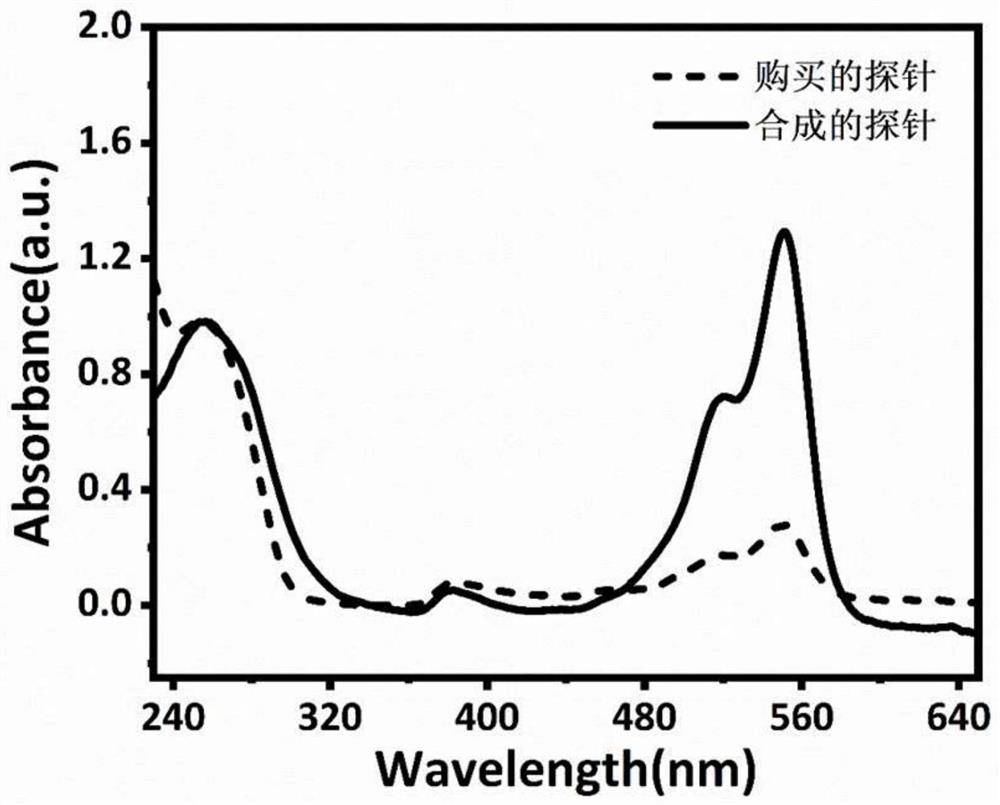
[0046] When the two probes were diluted to the same concentration of single-stranded DNA, both at 32ng / μL, the absorbance of cy3 at 550nm was measured by Nanodrop 2000, and the results were as follows figure 2 shown.
[0047] figure 2 The schematic diagram of the ultraviolet absorption spectrum shown is compared with the absorbance value of cy3 at 550nm, and the fluorescent molecule of the multi-fluorescent nucleic acid probe synthes...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com