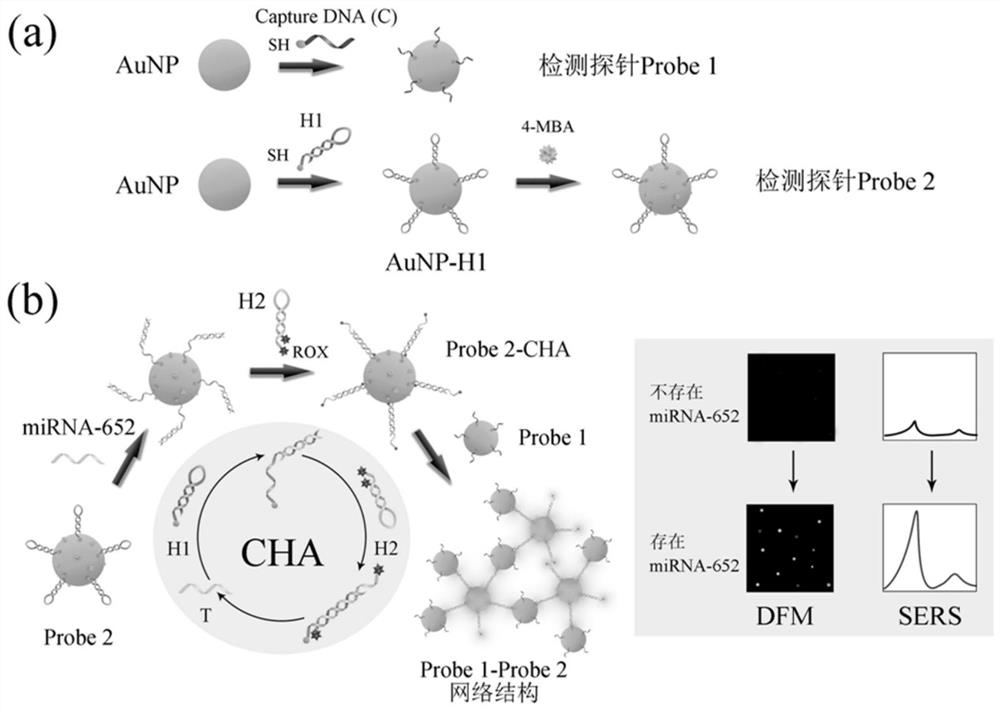
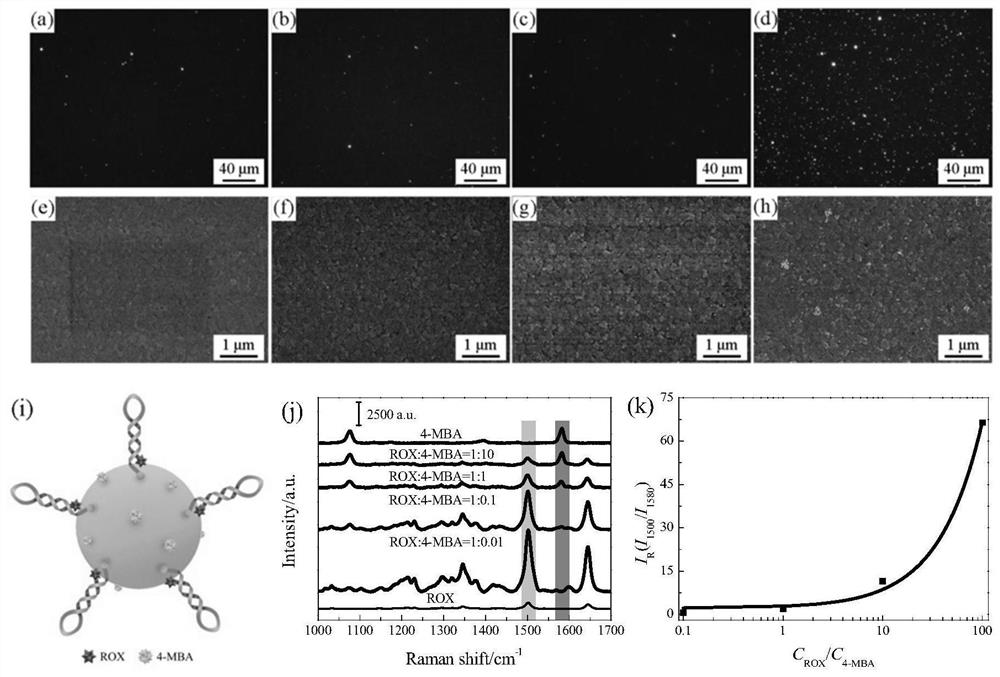
SPR-SERS dual-mode sensor for detecting disease nucleic acid markers as well as preparation method and application of SPR-SERS dual-mode sensor
A marker and sensor technology, applied in the fields of biological detection and spectroscopic detection, can solve the problems of missed diagnosis or diagnosis error, nucleic acid amplification, etc., and achieve the effects of reliable detection, good specificity and simple preparation process
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0060] Example 1 Preparation of SPR-SERS dual-mode sensor
[0061] 1. Preparation of the first reagent
[0062] (1) Place the hairpin DNA single strands H1 and H2 in a mixer for annealing at 95°C for 5 minutes to keep the hairpin structure;
[0063] (2) Mix 10 μL of 100 μM captured single-stranded C, 10 μL of 100 μM hairpin DNA single-stranded H1 with 10 μL of 100 mM TCEP solution (disulfide bond reducing agent) and place them in a constant temperature mixer at 25°C for overnight reaction To reduce disulfide bonds formed between sulfhydryl groups;
[0064] (3) Mix 100 μL 2.3nM AuNPs and 10 μL 10 μM captured single-stranded C in a 0.5×TBE solution, and incubate overnight at 25°C at 300 rpm to obtain a mixture 1;
[0065] (4) Add salt to the mixed solution 1 obtained in step (3) for aging: slowly add 10 μL of 2M NaCl solution to the above-mentioned mixture 1 in 4 times at intervals of 30 minutes, the final NaCl concentration is 0.3M, and then at 25°C 300rpm Shake overnight un...
Embodiment 2
[0076] Working curve and detection limit of embodiment 2SPR-SERS dual-mode sensor
[0077] The target miRNA-652 was diluted with 10% normal human serum to different concentrations of 0 (blank), 10 fM, 100 fM, 1 pM, 10 pM, 100 pM, 1 nM and 10 nM. Take 10 μL of the first reagent, 10 μL of the third reagent, and 5 μL of 10 μM of the second reagent and mix them in 45 μL of PBS buffer, add 5 μL of different concentrations of miRNA-652, shake in a mixer at 25°C for 3 hours, and centrifuge and wash 3 times. The final product was redispersed in 40 μL of PBS buffer for SPR-SERS dual-mode detection. SPR was detected by dark-field microscopy (DFM), and 20 μL of the final product after miRNA-652 detection was directly dropped on the center of clean indium tin oxide (ITO) glass (25 × 5 mm), adsorbed at 25 ° C for 1 min, and then washed with N 2 Blow dry, drop PBS buffer solution on the ITO glass and take DFM pictures. After the shooting was completed, Image Pro Plus software was used for...
Embodiment 3
[0079] Example 3 The specificity of SPR-SERS dual-mode sensor
[0080] The target miRNA-652 was diluted to 1pM with PBS buffer, and the complete mismatch sequence (UM), three-base mismatch sequence (TM) and single-base mismatch sequence (SM) were all diluted to 100pM with PBS buffer. Take 10 μL of the first reagent, 10 μL of the third reagent, and 5 μL of 10 μM of the second reagent and mix them in 45 μL of PBS buffer, add 5 μL of 1 pM miRNA-652, 100 pM complete mismatch sequence (UM), and 100 pM three-base mismatch sequence ( TM), 100pM single-base mismatch sequence (SM) and blank sample group (PBS buffer), after shaking in a mixer at 25°C for 3h, centrifugation and washing, SPR-SERS dual-mode detection was performed. Get the integral of the dark field colors corresponding to the target miRNA-652, complete mismatch sequence (UM), three-base mismatch sequence (TM), single-base mismatch sequence (SM) and blank sample group (PBS buffer) Optical density, SERS spectrum and the ra...
PUM
| Property | Measurement | Unit |
|---|---|---|
| particle diameter | aaaaa | aaaaa |
| diameter | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com