Tartary buckwheat sourced emodin glycosyltransferase as well as coding gene and application thereof
A technology of glycosyltransferase and emodin, applied in the direction of transferase, application, genetic engineering, etc., can solve the problems of unclear and blank metabolic mechanism
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0042] Example 1 Cloning of FtUGT74L2 CDS
[0043] The present embodiment adopts the method of gene cloning to clone and obtain the glycosyltransferase FtUGT74L2 gene from tartary buckwheat (Pinku No. 1) sterile seedling and carry out gene sequence information analysis, specifically as follows:
[0044] Select two-week-old seedlings of Pinku No. 1, take 50-100 mg of the seedlings, add liquid nitrogen to fully grind, and use the Trizol method to extract total RNA. Using this RNA as a template, use III 1st Strand cDNA Synthesis Kit (+gDNAwiper) kit (Nanjing Novizan Biotechnology Co., Ltd.) was used for reverse transcription to obtain the cDNA of the seedlings.
[0045] Design specific primers according to the ORF of FtUGT74L2:
[0046] FtUGT74L2-F: 5'-ATGAAGGTAGTTCCCGGG-3',
[0047] FtUGT74L2-R: 5'-AGCCCTTAACTCCTTCACAA-3';
[0048] PCR amplification was carried out using Pinku 1 cDNA as a template to obtain the CDS sequence of the target gene:
[0049] The PCR program was ...
Embodiment 2
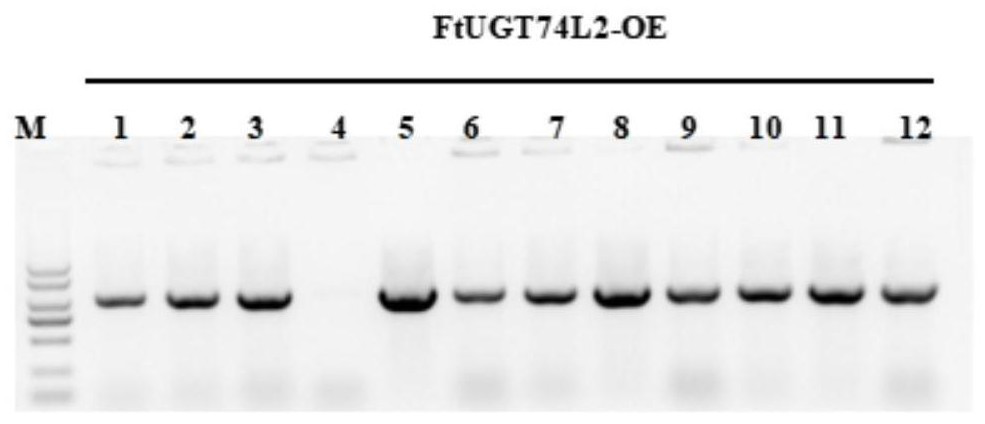
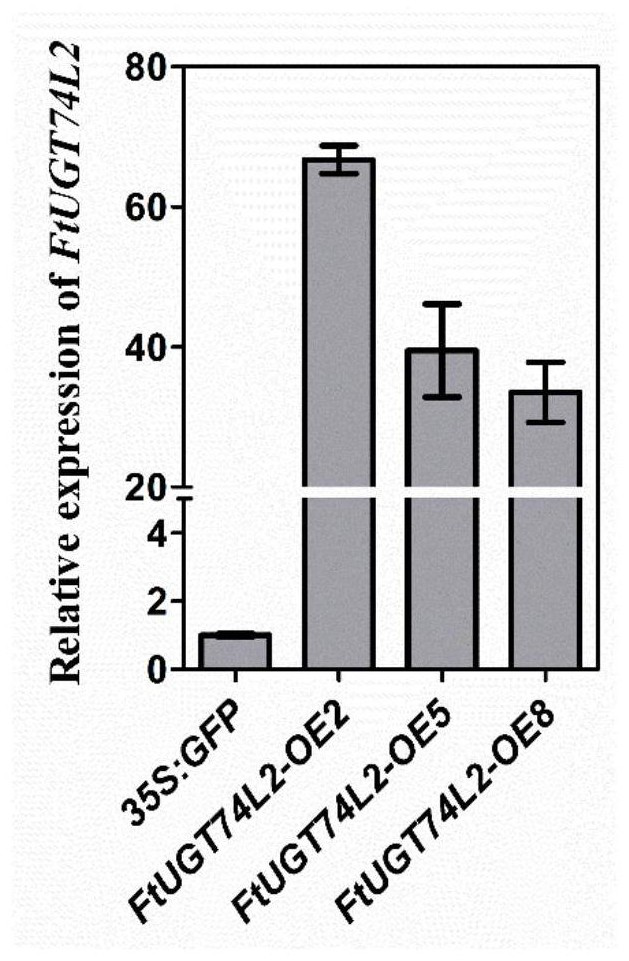
[0051] Example 2 Expression of FtUGT74L2 gene in tartary buckwheat hairy root and detection of expression level
[0052] The FtUGT74L2 gene is operably constructed in the expression control sequence to obtain the plant expression vector pCAMBIA 1302-FtUGT74L2 containing the FtUGT74L2 gene. Specifically, the construction method of the plant expression vector pCAMBIA 1302-FtUGT74L2 includes:
[0053] Homologous recombination primers were designed, and the full-length sequence of FtUGT74L2 was amplified by PCR using the FtUGT74L2-T vector as a template and OE-FtUGT74L2-F / R as primers.
[0054] Upstream primer: 1302-FtUGT74L2-NcoI-F:
[0055] 5'-ccatggATGAAGGTAGTTCCCGGG-3'
[0056] Downstream primer: 1302-FtUGT74L2-BglII-R:
[0057] 5'-agatctAGCCCTTAACTCCTTCACAA-3'.
[0058] After digestion, recovery and ligation transformation, the full-length sequence of FtUGT74L2 was inserted forward into the downstream of the CaMV35S promoter of the pCAMBIA-1302 vector, and the overexpressi...
Embodiment 3
[0063] Amino acid sequence analysis and comparison of embodiment 3 emodin glycosyltransferase
[0064] Blast the amino acid sequence of emodin glycosyltransferase FtUGT74L2 gene in NCBI database to obtain the GTs protein sequence of other species, use Bioxm software to translate the amino acid sequence, and predict its protein size and isoelectric point information.
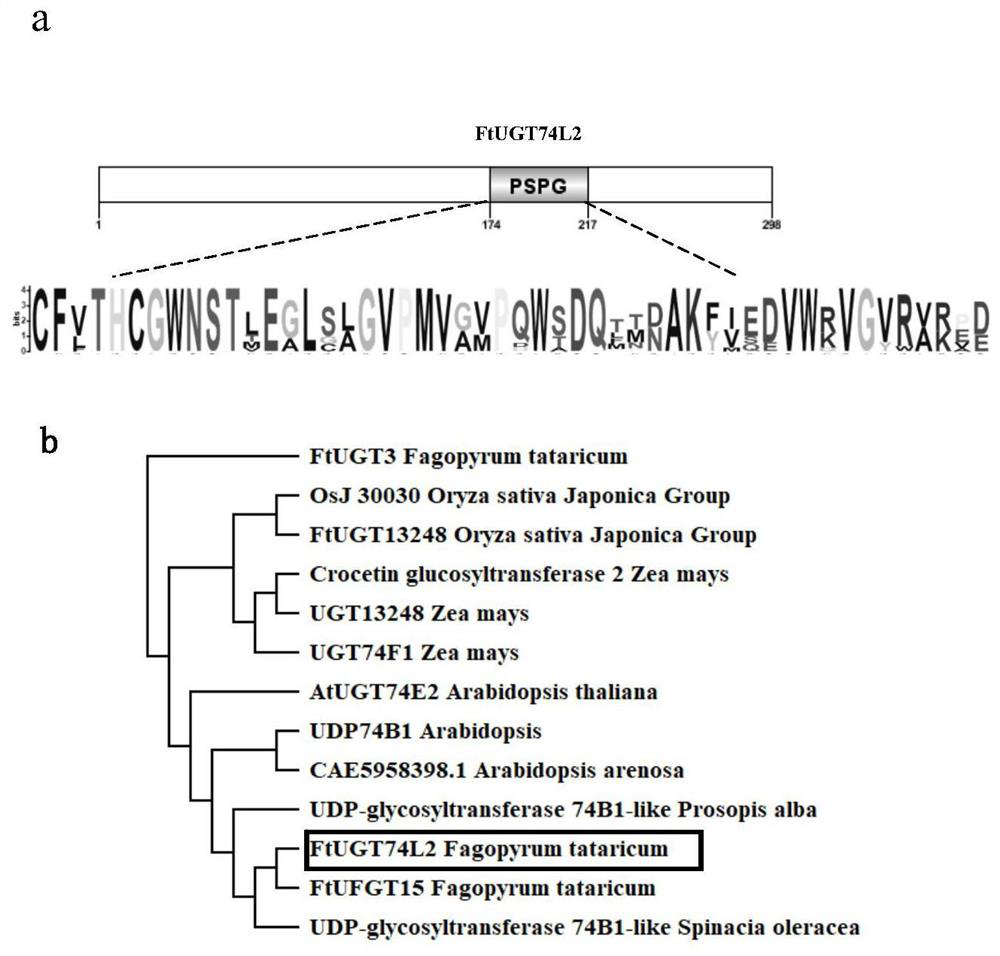
[0065]The length of the CDS of the gene of emodin glycosyltransferase FtUGT74L2 is 897bp (SEQ ID No.1), encoding 298 amino acids (SEQ ID No.2) with a protein molecular weight of 33.3KDa, and the isoelectric point (pI) of the encoded protein is 5.18 . Using NCBI's blast to analyze the conserved amino acid domain of FtUGT74L2 online, it was found that it has a glycosyltransferase (PSPG) domain, and the encoded protein belongs to the UGT-type glycosyltransferase ( image 3 a). The UGT protein sequences of other species were obtained by Blast comparison in the NCBI database, and the clustering tree was constructed ...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Molecular weight | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com