Detection method of target nucleic acid and application thereof
A detection method and target nucleic acid technology, applied in the field of target nucleic acid detection, can solve problems such as prolonging detection time
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0044] Embodiment 1: the preparation of albumen
[0045] This embodiment relates to 3 kinds of plasmids of the III-A type CRISPR system, as follows:
[0046] The first method: using the pCDF plasmid vector as a template, five subunit proteins (including TtCsm1, TtCsm2, TtCsm3, TtCsm4, and TtCsm5) in the TtCsm complex are fused and expressed, and site-directed mutation is carried out, the 34th amino acid of Csm3 is asparagus Amino acid D is mutated to alanine A (D34A) to inactivate target RNA cleavage mediated by Csm3.
[0047] The second: using pACYC as a plasmid vector, inserting TtCsm1 (Cas10) protein and an artificially designed CRISPR array, the synthetic CRISPR array can recognize the N gene of SARS-CoV-2.
[0048] The third method: use pC0075 as a plasmid vector, insert the TtCsm6 protein sequence and fuse the SUMO tag to facilitate purification.
[0049] Co-transform Escherichia coli BL21(DE3) with the first plasmid and the second plasmid, grow in LB broth at 37°C to ...
Embodiment 2
[0051] Example 2: Preparation of fluorescent chip
[0052] This embodiment provides a glass slide anchored with a fluorescent reporter group, and the specific steps are as follows:
[0053] (1) Cover the surface of coverslip (24×30mm) with biotin through chemical modification: first, wash the slide, etch with acetone for 30min, then corrode with potassium hydroxide for 1h, rinse with distilled water Then dry under nitrogen; soak in aminosilane solution for 10 minutes in a dark environment, sonicate for 1-2 minutes and then return to a dark environment for 10 minutes; add biotin to the surface of the slide in bicarbonate buffer and incubate overnight ; Store at -80°C.
[0054] (2) Add streptavidin (streptavidin) to combine with biotin: use tape cut into thin strips on the surface of the slide to form a loading area, add streptavidin to the surface of the slide and incubate for 10 min, Residual streptavidin not bound to biotin was rinsed with distilled water.
[0055] (3) Anc...
Embodiment 3
[0056] Embodiment 3: the detection of target nucleic acid
[0057] This embodiment provides a method for detecting target nucleic acids for non-disease diagnosis purposes, comprising the following steps:
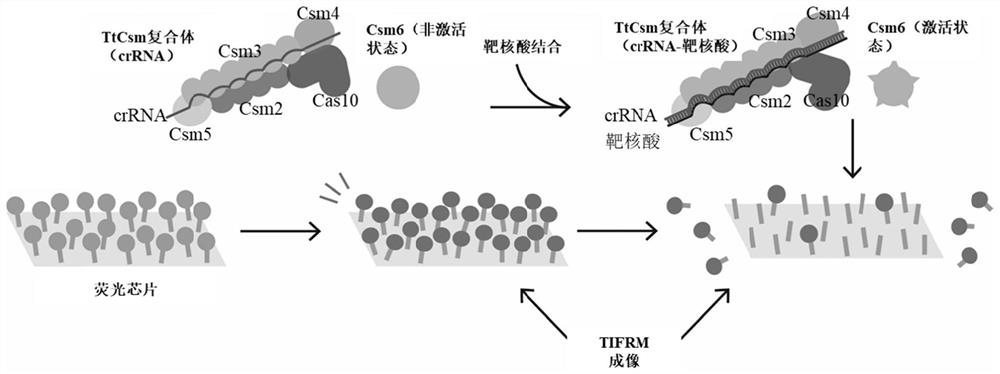
[0058] (1) Before the TtCsm complex, TtCsm6 protein and other reaction systems in Example 1 are added to the fluorescent chip slides prepared in Example 2, first use TIRFM to perform fluorescence excitation and imaging on the slides, and record the initial fluorescence intensity imaging.
[0059] (2) Add 250μM ATP, 500nM TtCsm complex, 2500nM TtCsm6 protein, target nucleic acid sample and reaction buffer (20mM Tris-HCl, pH 7.9, 200mM Monopotassium Glutamate, 10mM Ammonium Sulfate, 5mM Magnesium Sulfate , 1mM TCEP) and incubated for 5min, washed the sheared fluorescent reporter gene with distilled water and imaged again with TIRFM to obtain the fluorescence intensity after the reaction. concentration of target nucleic acid.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com