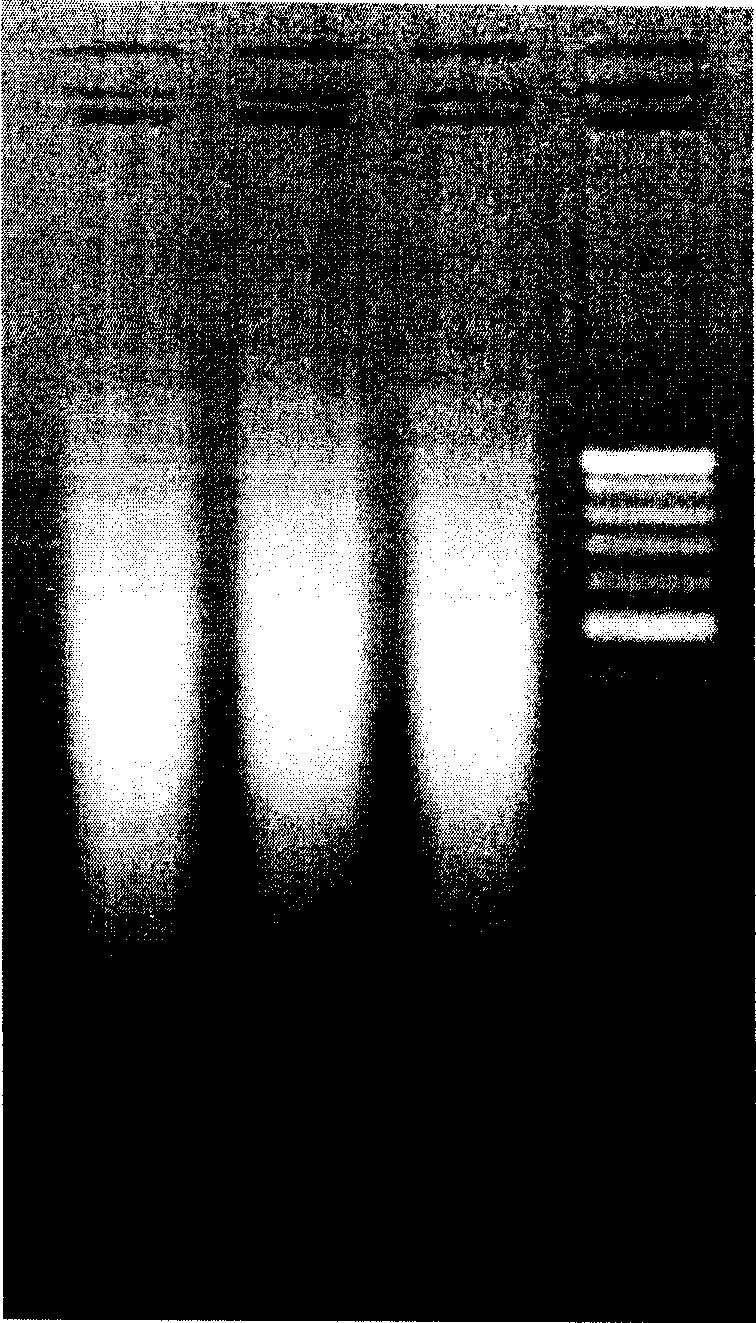Test chipe of cytochrome P450 gene hereditary variation and its application
A technology of cytochrome and detection chip, applied in the field of detection chip of genetic variation of cytochrome P450 gene, which can solve the problem of lack of functional variation
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0078] Example 1 Preparation of gene chip
[0079] 1. Probe Dissolution
[0080] Each probe was diluted with TE solution to a final concentration of 10 mM. Mix the probe with a concentration of 10mM and the PBS solution with a concentration of 200mM in a medium volume of a 384-well plate, seal the 384-well plate with an adhesive sheet, shake at room temperature for 2 minutes, centrifuge, and store at -20°C for sample application .
[0081] 2. Spotting
[0082] The pre-designed and synthesized probes are loaded onto the solid-phase carrier substrates made of glass slides, silicon wafers, etc. through contact spotting or inkjet spotting. The film base adopts Cell Associates CSS-100 aldehyde base film base, and the Ominigrid 100 model spotting instrument of GeneMachine Company is applied at a humidity of 65-75% (based on the FullMoon film base) and a temperature of 25°C. The format of spotting is 1×3, and each matrix is 8×18. After the spotting is completed, leave the chip ...
Embodiment 2
[0083] Embodiment 2 Processing and labeling of samples to be tested
[0084] 1. Human genomic DNA extraction and target gene amplification
[0085] Using the FlexiGene DNA Kit (250) (QIAGEN, Cat.No.51206) kit to extract genomic DNA from human peripheral blood, the specific steps are as follows:
[0086] (1) Mix ACD (citric acid 8g / L, sodium citrate 22g / L, glucose 24.5g / L) anticoagulated peripheral whole blood, take 1ml into a 15ml centrifuge tube, add 2.5ml buffer FG1, fully Mix upside down;
[0087] (2) Centrifuge at 2000 g for 5 minutes, discard the supernatant. Let stand for 2 minutes;
[0088] (3) Add 500ul buffer FG2 / QIAGEN protease mixed reagent, mix immediately until the original precipitate completely disappears;
[0089] (4) Water bath at 65°C for 10 minutes, shake at a constant speed at 40 times / min;
[0090] (5) Add 500ul 100% isopropanol and mix thoroughly until visible flocculent DNA precipitates;
[0091] (6) Centrifuge at 3000g for 5 minutes, discard the s...
Embodiment 3
[0114] Example 3 Hybridization, washing and result detection
[0115] Fluorescence-labeled PCR products were denatured at 95°C for 10 minutes, and immediately placed on ice for hybridization. The hybridization reaction system included:
[0116]
[0117] The reaction conditions were incubation at 48°C for 120 min, followed by 1× wash buffer I (5×SSC, 0.1% SDS), 1× wash buffer II (2×SSC, 0.1% SDS) and 1× wash buffer III ( 1×SSC) at 42°C for 10 min each, and finally with ddH2O for 0.5 min.
[0118] After washing, the chips were dried and scanned with a GenePix 4000B confocal laser scanner (other laser scanners can also be used). The hybridization results obtained by scanning the hybridized chips are as follows: Figure 4 As shown, then use GenePix Pro to process the image to obtain the data file, and then analyze the data file to obtain the genotype of the target gene. Figure 4 The typing results shown are CYP2D6*10, CYP2C19*1, CYP2C9*1 and CYP3A5*3.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com