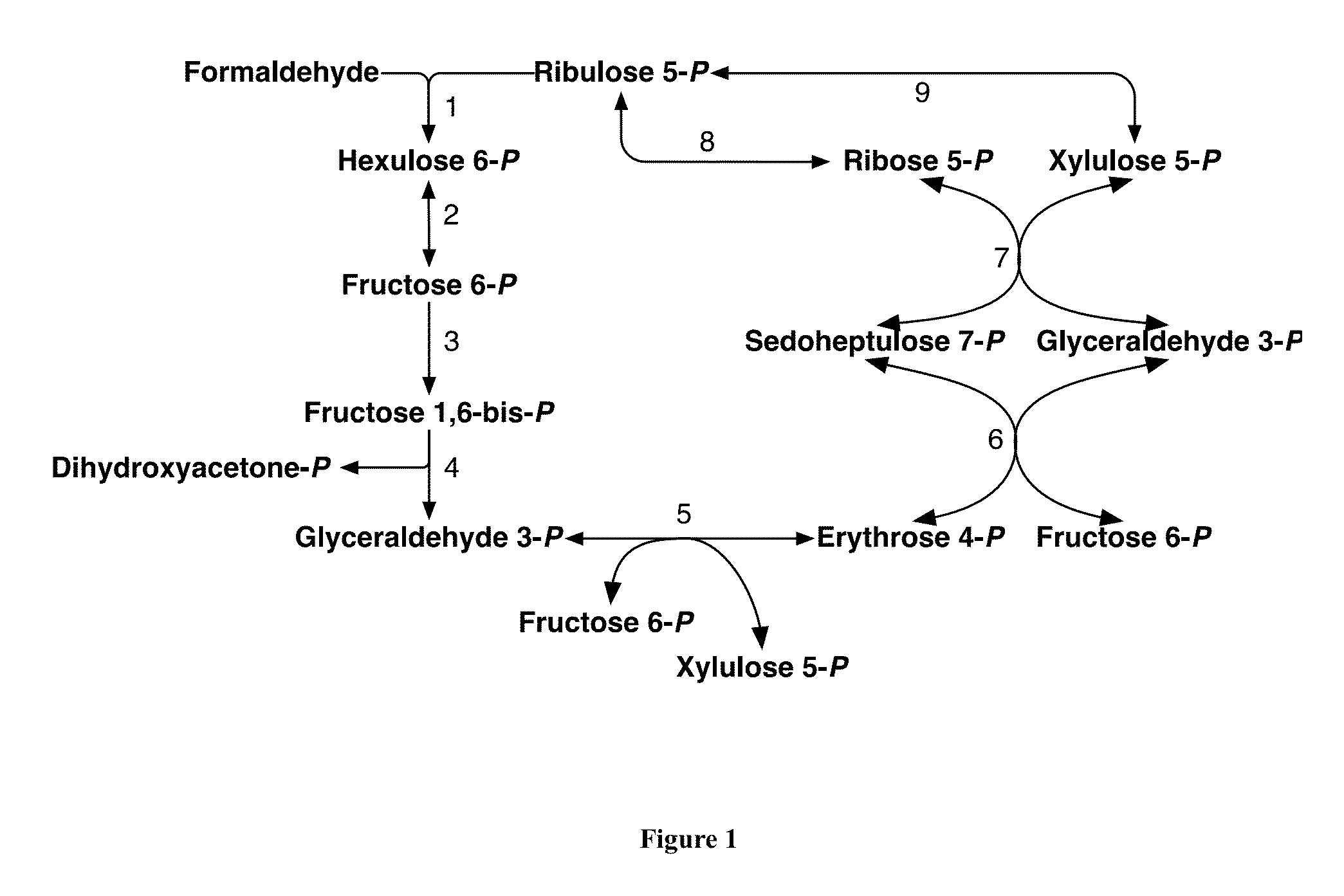
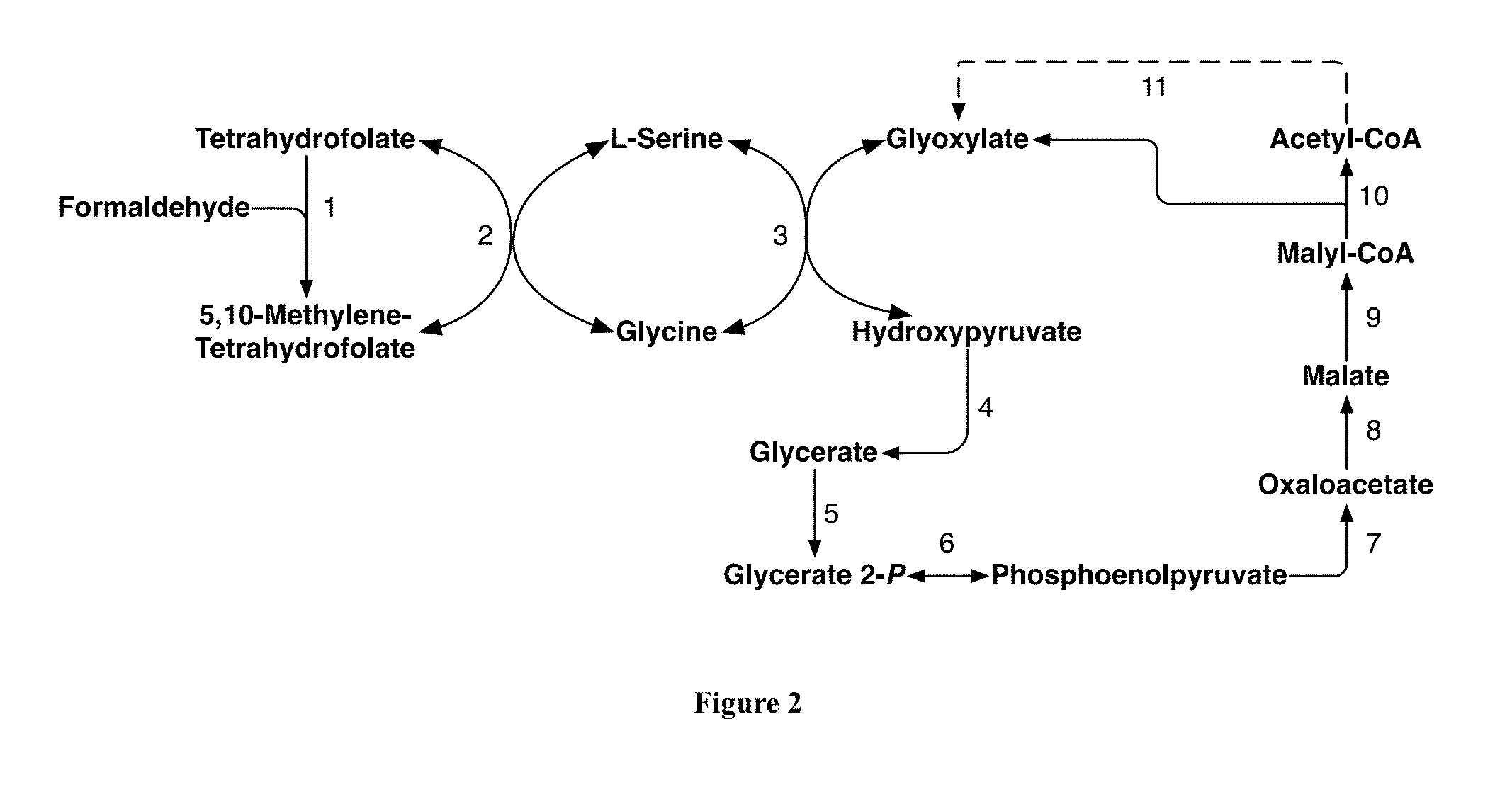
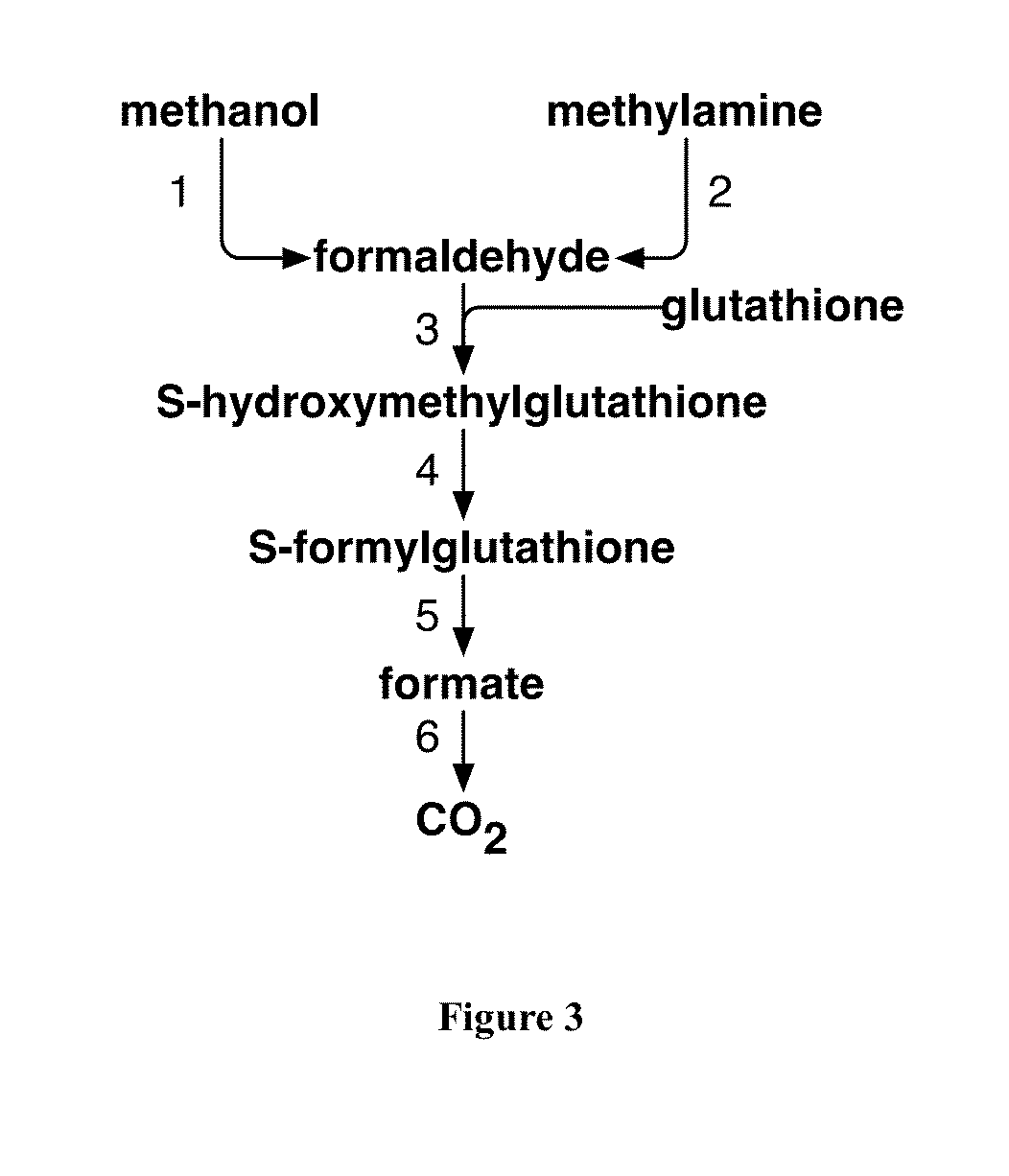
Methods and Systems for Methylotrophic Production of Organic Compounds
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
Optimization of Growth Medium for Paracoccus sp. when Using Formate as the C1 Compound
[0257]Paracoccus zeaxanthinifaciens ATCC 21588, Paracoccus versutus ATCC 25364, and Paracoccus denitrificans ATCC 13534 were obtained from the American Type Culture Collection (ATCC).
[0258]Strains were tested for the ability to grow aerobically on sodium formate as a sole source of carbon and / or energy using MOPS minimal medium (Teknova, Inc.) with sodium formate as a sole carbon source at 37C. Unlike previous media used to evaluate the formate-dependent growth of Paracoccus, this medium contains defined levels of trace elements molybdenum, boron, copper, zinc, manganese, and other trace metals.
[0259]Growth was conducted in various high-throughput machinery capable of monitoring growth by light scattering at 600 nm, including a Gemini SpectraNax plate reader (Molecular Devices, Inc.), a Tecan M3000 plate reader (Tecan, Inc.), and a BioLector device (m2p-labs, Inc.). For the BioLector, the CO2 gas c...
example 2
Automatable Protocol for Conjugative Transfer of Plasmids from E. coli Donors to Paracoccus sp
[0267]E. coli strain 517-1 was obtained from the Yale E. coli Genetic Stock Center. Paracoccus denarificans PD1222 was obtained from Stephen Spiro (University of Texas at Dallas). E. coli S17-1 strain is tra+, meaning it is able to mobilize for conjugative transfer those plasmids harboring a mob+genotype. Plasmid pDIY313K, obtained from Dariusz Bartosik (University of Warsaw, Poland) and described by his laboratory [J Microbiol Methods, 2011, 86(2):166-74, DOI: 10.1016 / j.mimet.2011.04.016], and introduced into E. coli 517-1 by standard methods.
[0268]E. coli S17-1 was grown on Luria broth with carbenicillin overnight. Paracoccus versutus ATCC 25364 was grown overnight on MOPS minimal medium (Teknova, Inc.) with 50 mM sucrose and 40 mM sodium nitrate.
[0269]The next day, E. coli S17-1 was subcultured in antibiotic-free Luria broth for >4 hr. Paracoccus strains were subcultured in identical MOP...
example 3
Genome Sequencing of Paracoccus Strains
[0272]Genomic DNA was isolated from Paracoccus zeaxanthinifaciens ATCC 21588, Paracoccus versutus ATCC 25364, and Paracoccus denitrificans ATCC 13534 using a Wizard Genomic DNA Isolation Kit (Promega, Inc.). The resulting DNA samples were fragmented and converted to paired-end libraries for whole-genome shotgun sequencing on a 454 pyrosequencing platform (Roche, Inc.).
[0273]For Paracoccus denitrificans, 37,585,886 paired reads, each 100 nt in length, were obtained. This represents approximately 3.7 gigabases of sequence data, or approximately 730-fold coverage of the 5.2 megabase genome of Paracoccus denitrificans PD1222 (Genbank accession numbers CP000489, CP000490, and CP000491 for chromosome 1, chromosome 2, and a 653,815 bp megaplasmid, respectively). Reads were assembled first by de-novo assembly and second by mapping the de-novo contigs to the published PD1222 genome.
[0274]The resulting reads could be assembled into a crude whole-genome a...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Temperature | aaaaa | aaaaa |
| Temperature | aaaaa | aaaaa |
| Time | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com