Gene sequence and amino acid constitution of (D)-specific carbonyl reduction enzyme of Morganella morganii proteus
A technology of carbonyl reductase and proteus, applied in the field of bioengineering, can solve the problems of blank genome sequence and few studies
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0016] Example 1: The (D)-specific carbonyl reductase of M. morganii CMCC (B) 49208 was obtained by conventional protein separation and purification methods. Take a certain amount of enzyme solution and add 0.2mol / L pH 7.5 phosphate buffer, 150mg / L MAK, 100μmol / L glucose, 10mol / L NADH, 15U / mL glucose dehydrogenase, make up to 1mL with double distilled water, and incubate at 30℃ Reaction for 10 hours, HPLC (Agilent 1100 serices chromatograph, ZORBAX SB C18 column, G1314A ultraviolet detector, column temperature: 40°C, detection wavelength: 220nm, injection volume: 10μL, mobile phase: methanol: 0.02mol / L diphosphate Potassium hydrogen buffer: acetic acid: triethylamine=4:96:0.2:0.13 (v:v:v:v)). The detection result shows that more than 89% of the MAK in the reaction solution is converted into d-pseudoephedrine.
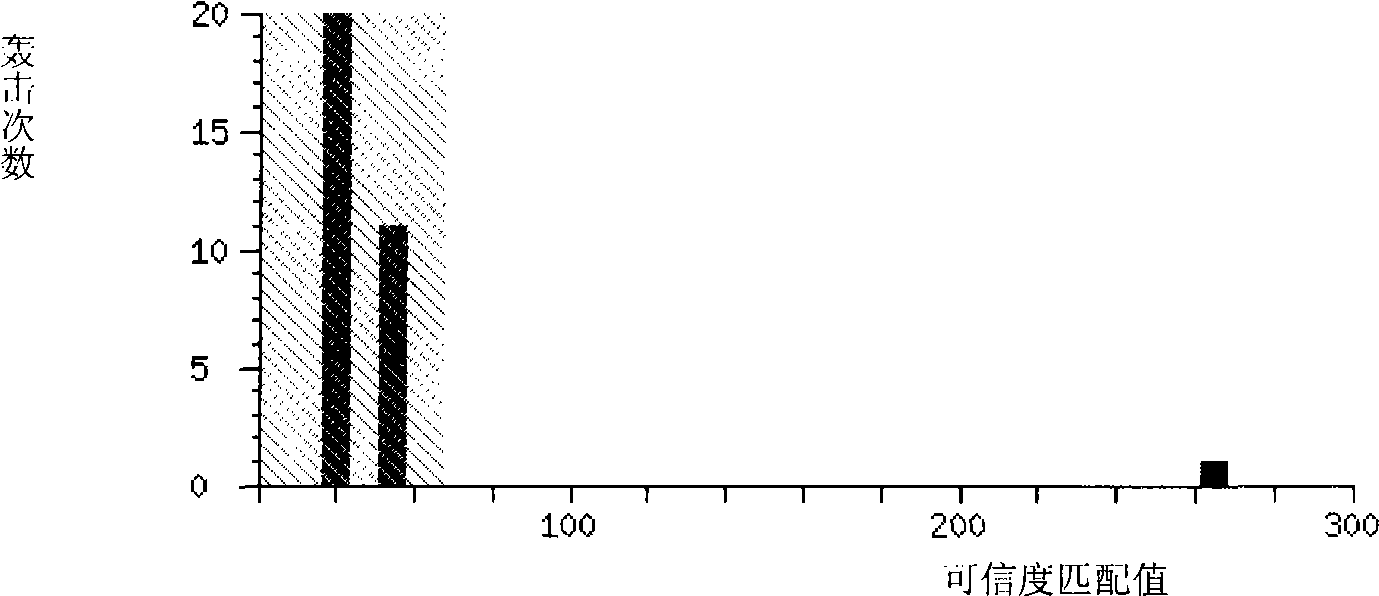
[0017] Using the MADLI-TOF-MS method to obtain the peptide fingerprint of the carbonyl asymmetric reductase, such as figure 1 shown. NCBI used MASCOT to identify the...
Embodiment 2
[0028] Embodiment 2: Get the upper and lower two parts ("IAIHDTTL" and "IATYVAAD") nucleotide sequence design and merge primers:
[0029] The upstream primer Z1 is: 5'-ATHGCNATHCAYGAYACNAC-3';
[0030] The downstream primer Z1 is: 5'-GCNGCNACRTANGTNGCDAT-3'
[0031] PCR amplification was performed on the bacterial genome of M. morganii CMCC(B)49208. Reaction conditions: pre-denaturation at 95°C for 5 minutes, denaturation at 95°C for 30 seconds, annealing at 53°C for 45 seconds, extension at 72°C for 1 minute, and extension at 72°C for 10 minutes after 30 cycles to obtain a gene fragment of about 900 bp. Reclaim the specificity fragment about 900bp, determine this gene fragment sequence, after this section sequence is compared with the database published above the NCBI website, obtain the leucine dehydrogenase gene ( leucine dehydrogenase LeuDH gene).
[0032] Several gene sequences with high homology were selected and their conserved sequences were analyzed to design prime...
Embodiment 3
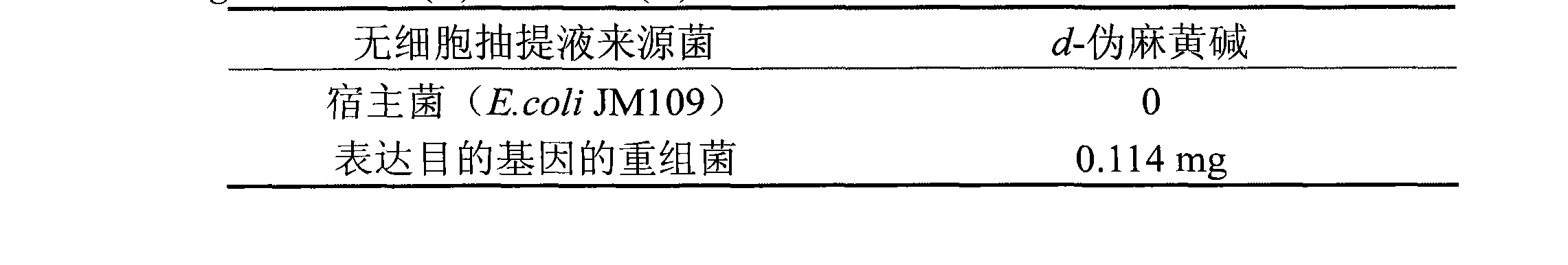
[0039] Example 3: After the extracted plasmid pKK223 was digested with PstI, it was ligated with the same digested target gene fragment and then transformed into E.coli JM109 competent cells, coated with LB plates containing ampicillin, randomly picked transformants, and extracted Recombinant plasmid, PstI enzyme digestion to verify that the recombinant plasmid inserted into the target gene is then digested with EcoRI to verify that the recombinant plasmid with the forward connection of the target gene is obtained, and the recombinant bacteria containing the recombinant plasmid are further verified for enzyme activity.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com