Jatropha curcas farnesyl pyrophosphate synthase protein encoding sequence and application thereof in plants
A Jatropha curcas, nucleotide sequence technology, applied in the fields of genetic engineering and molecular biology
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0137] Cloning of JcFPS Protein Gene of Jatropha curcas
[0138] 1. Tissue separation (isolation)
[0139] The seeds of Jatropha curcas come from Panzhihua, Sichuan Province. After the seeds of Jatropha curcas are collected, they are stored in the laboratory.
[0140] 2. RNA isolation (RNA isolation)
[0141] Peel off the seed coat of Jatropha curcas seeds, take out the seed kernels, grind them with a mortar, add liquid nitrogen, grind them into powder, take 100mg and transfer them to 1.5mL EP tubes, and extract total RNA (two-step lysis method) . The quality of total RNA was identified by formaldehyde denaturing gel electrophoresis, and then the RNA content was determined on a spectrophotometer.
[0142] 3. Cloning of Full-length cDNA
[0143] According to the amino acid conserved sequence of some plant FPS, design annexation primers, use the principle of homologous gene cloning, and use the SMARTTMRACE cDNA amplification method (Clonetech kit) to carry out full-length cD...
Embodiment 2
[0153] Sequence Information and Homology Analysis of JcFPS Protein Gene of Jatropha curcas
[0154] The length of the full-length cDNA of the new Jatropha curcas JcFPS protein of the present invention is 1029bp, and the detailed sequence is shown in SEQ ID NO.1, wherein the open reading frame is located at nucleotides 221-1246. The amino acid sequence of Jatropha curcas JcFPS protein was deduced according to the full-length cDNA, with a total of 381 amino acid residues, a molecular weight of 43784.45, and a pI of 5.54. See SEQ ID NO.2 for the detailed sequence.
[0155] The full-length cDNA sequence of the Jatropha curcas JcFPS protein and its encoded protein were nucleotide aligned in the Non-redundant GenBank+EMBL+DDBJ+PDB and Non-redundant GenBank CDStranslations+PDB+SwissProt+Superdate+PIR databases using the BLAST program. Protein homology search, it was found that it has 87% homology (attached table 2) with Brasiliensis HbFPS gene (AB294712) at the nucleotide level; AA...
Embodiment 3
[0156] The structural characteristic analysis of embodiment 3 Jatropha curcas FPS protein
[0157] 1. Domain analysis of Jatropha curcas FPS protein
[0158] The amino acid sequence of the Jatropha curcas FPS protein is retrieved in the NCBI database (URL: http: / / www.ncbi.nlm.nih.gov / Blast.cgi), and the results show that:
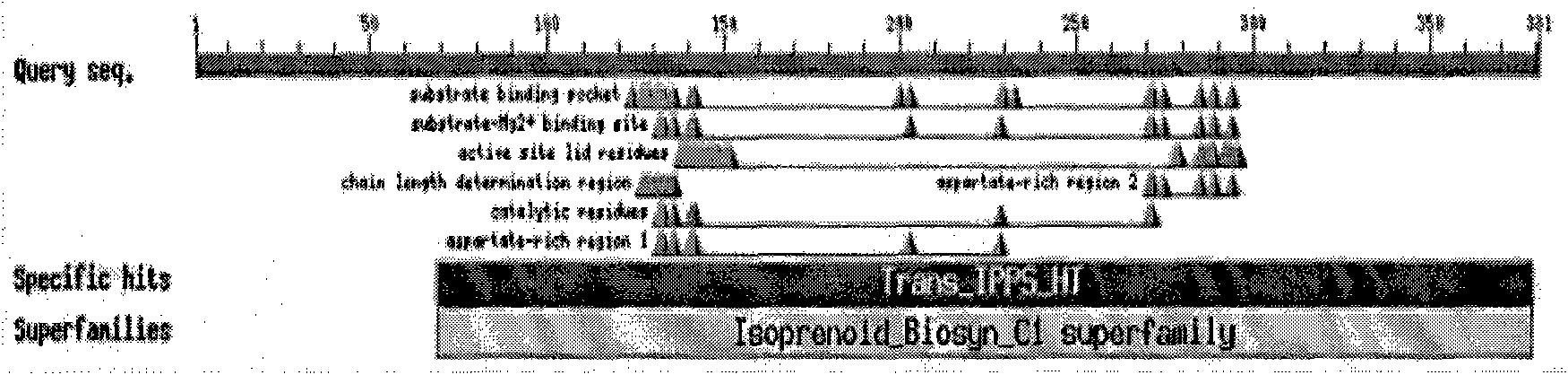
[0159]In the amino acid sequence, there is the catalytic site of Trans-Isoprenyl Diphosphate Synthases (Trans-Isoprenyl Diphosphate Synthases, Trans IPPS) (ie, amino acid 69-303 from the amino terminal (N terminal) of SEQ ID.1 in the sequence listing Residues.
[0160] 2. Hydrophobicity analysis of Jatropha curcas FPS protein
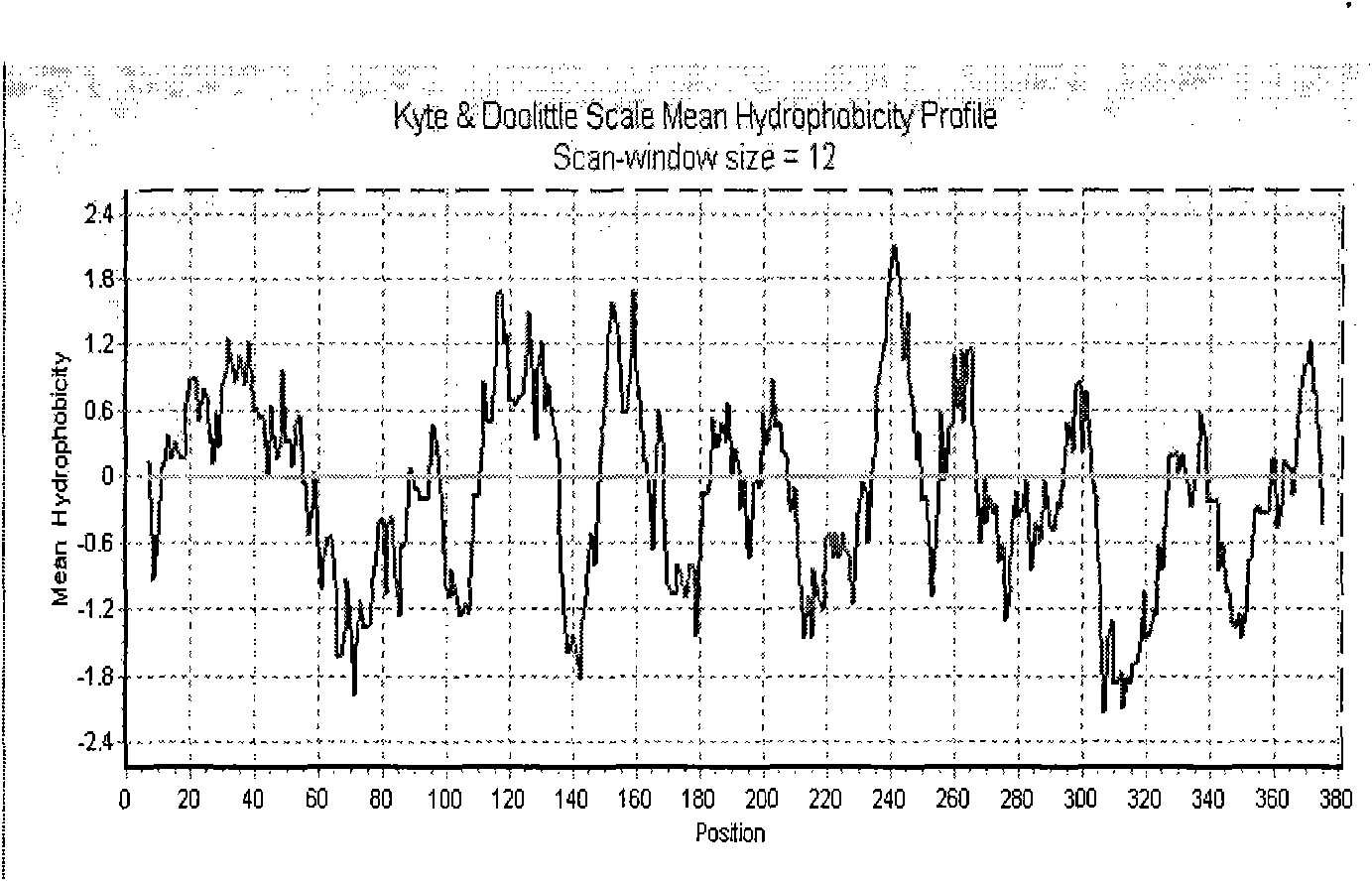
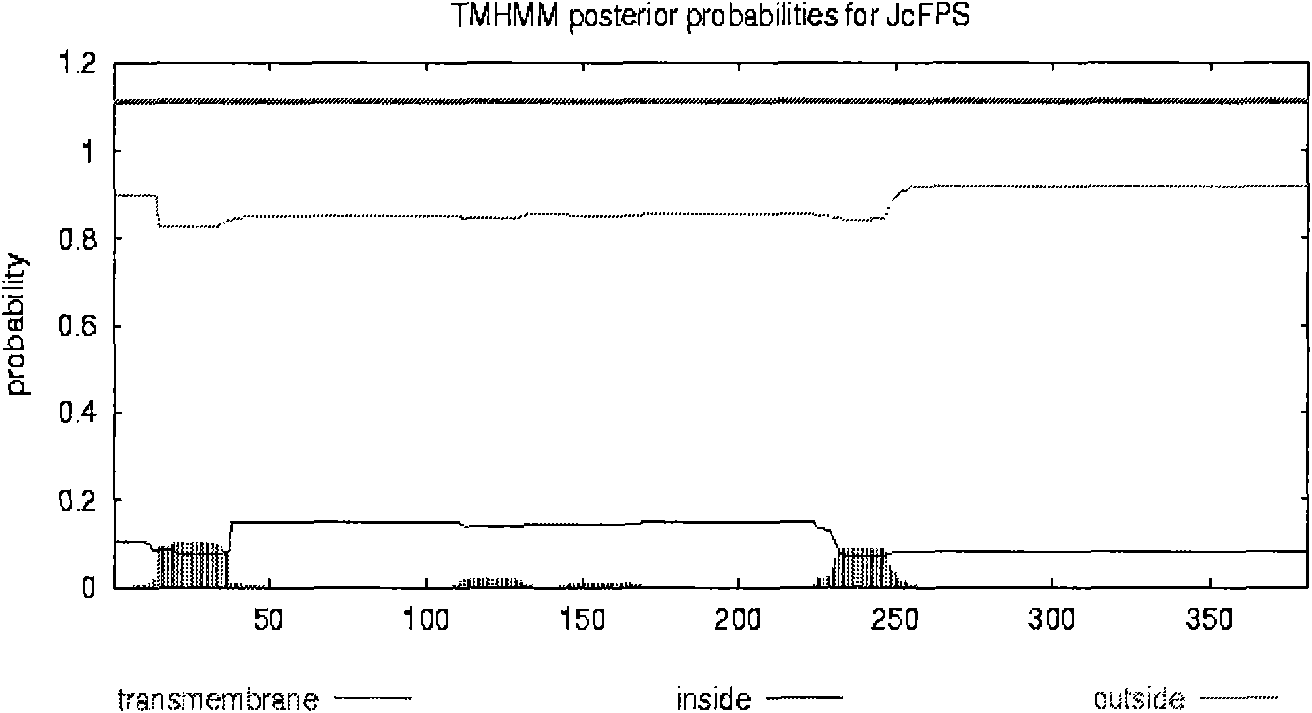
[0161] Analysis of the mature FPS after shearing of the transit peptide shows that, on the whole, the hydrophilic amino acids are evenly distributed in the entire peptide chain, and the entire polypeptide chain is hydrophilic without obvious hydrophobic regions. The hydrophobicity analysis chart of the present invention is establi...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com