Separation and purification method of wild-type p53 protein
A separation and purification, wild-type technology, applied in the field of wild-type p53 protein separation and purification, to achieve the effect of improving yield and maintaining activity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1p5
[0021] The separation and purification of embodiment 1p53 protein
[0022] 1. Materials and methods
[0023] 1. Materials and Reagents
[0024] 1.1 Materials: human liver cDNA, DH5α strain, JM109 competent bacteria, H1299 cells were preserved by our laboratory; eukaryotic expression vector pCMV-Myc, mouse (mouse) anti-p53 monoclonal antibody was purchased from Clontech Company; PCR amplification kit; PCR product purification kit was purchased from Boehringer Company; cell culture medium and fetal bovine serum were purchased from Hangzhou Sijiqing Bioengineering Materials Company; cells were transiently transfected with Invitrogen liposome transfection kit; p53 monoclonal antibody (DO1) was purchased from In SANTACRUZ Company: All primers used were synthesized by Shanghai Boya Biotechnology Co., Ltd.
[0025] 1.2 Reagents:
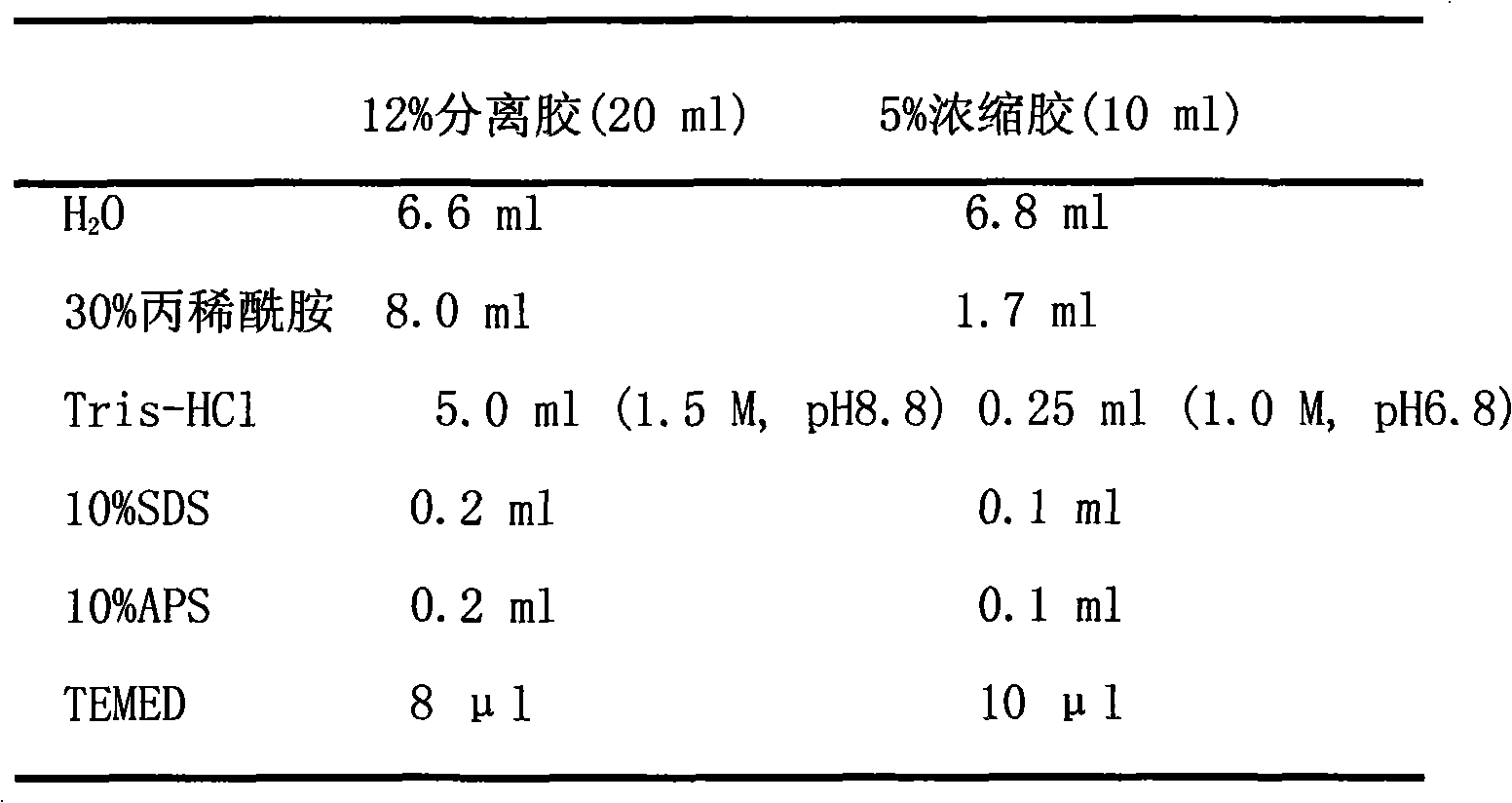
[0026] 50×TAE: Dissolve 242g of Tris base in 600ml of ddH2O, add 57.1ml of glacial acetic acid, 100ml of 0.5M EDTA (pH 8.0), and dilute to 1L.
[0027]...
Embodiment 2
[0072] Example 2 The effect of p53 recombinant protein on H1299 tumor cell apoptosis measured by flow cytometry
[0073]H1299 cells induced by p53 recombinant protein at a concentration of 180ng / ul were selected, and the cells were collected at four time points of 12h, 24h, 48h, and 72h, washed once with 1×PBS (pH7.4), and placed in citrate buffer After 1 h, centrifuge to remove the supernatant. Appropriate amounts of trypsin digestion solution and trypsin inhibitor were added successively, and PI was added to stain in the dark for 15 minutes, and then a single cell suspension was formed for detection on the machine. The H1299 tumor cells treated with p53 recombinant protein at four time gradients can detect apoptosis peaks in flow cytometry, and the apoptosis rate is greatly improved compared with the control group. It can be seen that with the prolongation of the action time of P53 recombinant protein, the apoptosis rate increased significantly. The apoptotic rate of the e...
Embodiment 3
[0074] Example 3 Using the BIACORE molecular interaction instrument to detect the interaction between the purified p53 protein and small molecules
[0075] BIACORE molecular interaction instrument is based on surface plasmon resonance technology to track the interaction between biomolecules. Since it does not require any markers and truly reflects the various interactions between biomolecules, it has been widely used by researchers in recent years. BIACORE molecular interaction instrument was used to detect the interaction between p53 recombinant protein and Hbx. The concentration of recombinant protein is determined in advance, and protein coupling can be carried out when a certain concentration requirement is reached. Dilute the target protein to be analyzed to 50ug / ml with sodium acetate buffer solution of pH 4.0, 4.5, 5.0 and 5.5, respectively, and then inject the sample, so that the above protein dilutions flow through the surface of the CM5 sensor chip successively. Th...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com