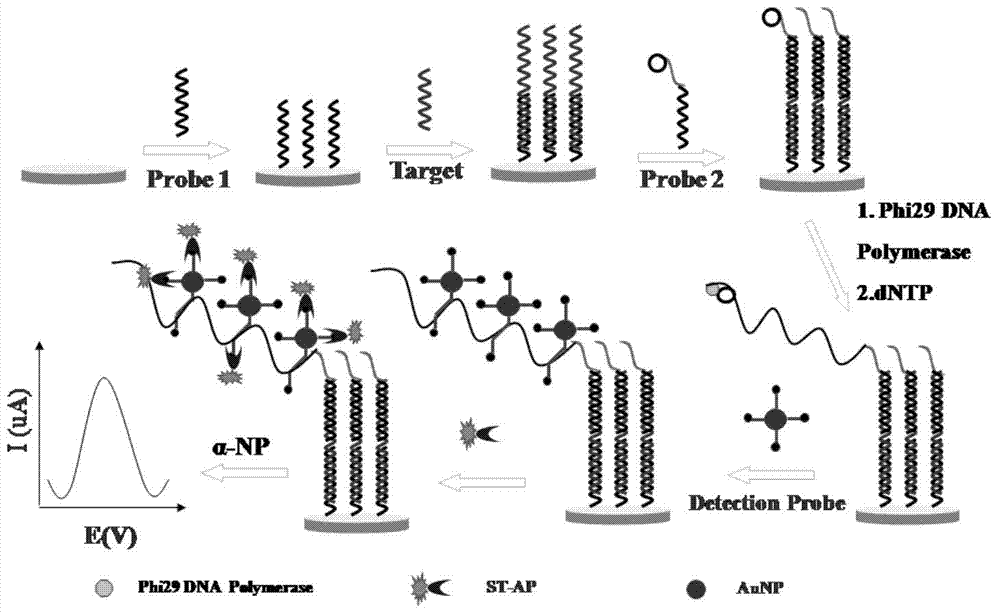
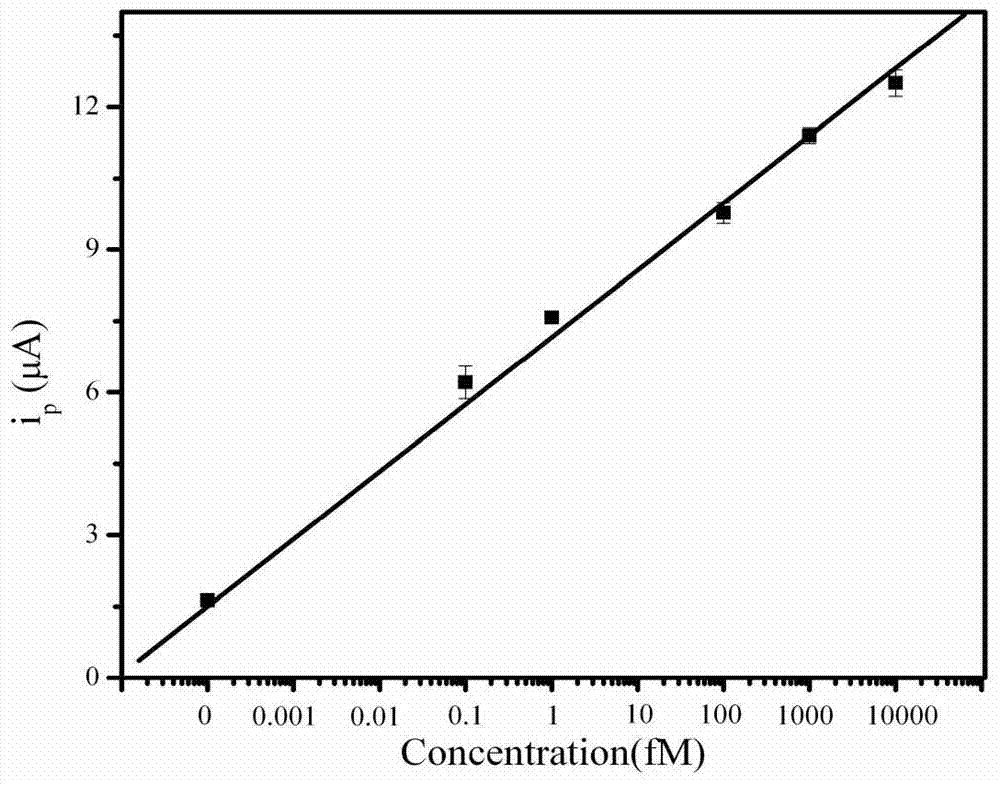
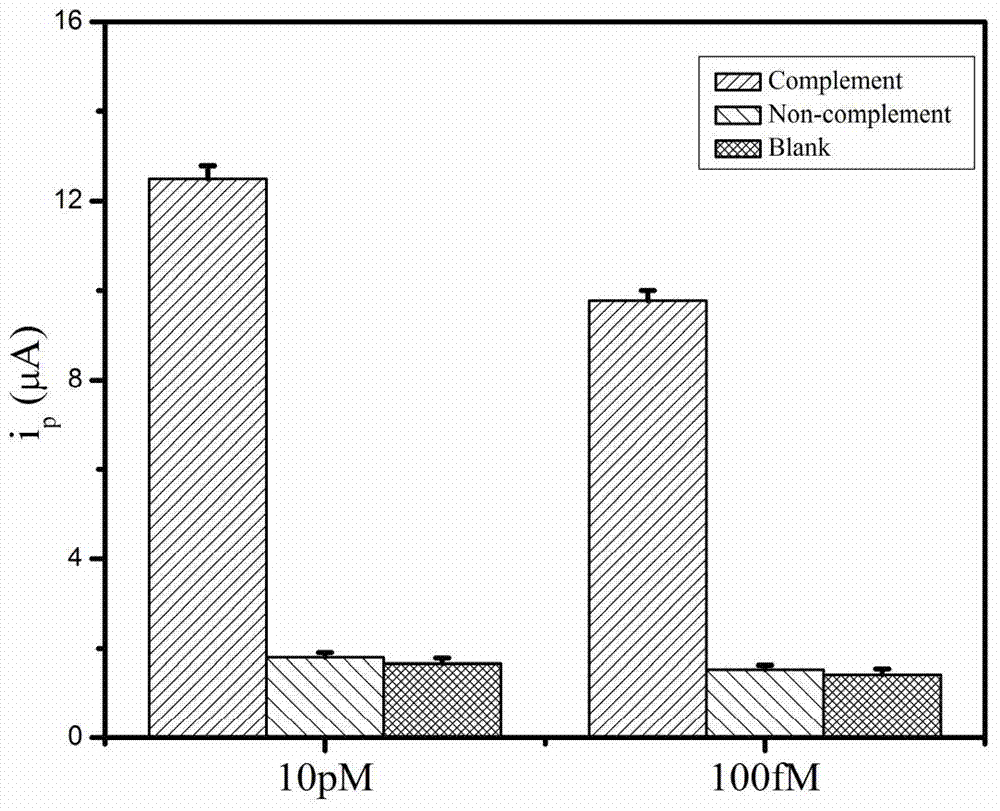
Method for detecting salmonella invA gene based on rolling circle amplification and gold nanoparticles
A technology of rolling circle amplification and gene detection, which is applied in the detection field of Salmonella gene, can solve the problems of high cost of nanoparticles and fluorescent probes, and achieve the effects of low detection cost, improved sensitivity and high sensitivity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0031] Embodiment 1. Preparation of circularized DNA
[0032] Mix 100nmol 5' end phosphorylated template DNA to be circularized with 100nmol probe 2 in 100μL ligation buffer (pH7.5, 50mM Tris, 10mM MgCl 2 , 10mM dithiothreitol, 0.5mM ATP), then add 0.2U of T4DNA ligase, ligation reaction at 37°C for 60 minutes; inactivate T4DNA ligase at 65°C for 10 minutes, and the circularized template DNA obtained at -20°C Save it for future use. The 5' end phosphorylated template DNA sequence to be circularized is: 5'-p-CTCAGCTGTGTA ACAACATGAAGATTGTAGGTCAGAACTCACCTGTTAGAAACTGTGAAGAT CGCTTATTA TGTCCTATC-3', and the sequence of probe 2 is: 5'-AATACTCATCTGTTTACCG GGCATAAAAAAAAAACACAGCTGAGGATAGGACAT-3'.
Embodiment 2
[0033] Embodiment 2, preparation of gold nanoparticles and gold nanoprobes
[0034] (1) Soak all utensils in aqua regia for 24 hours, rinse them with double distilled water and set aside.
[0035] (2) Prepare 100mL of 0.01% HAuCl 4 4H 2 O deionized aqueous solution, placed on a heating magnetic stirrer and stirred, after the solution boiled, quickly added 4mL of 1% trisodium citrate solution. The color of the gold nanoparticle solution turns black at first, and then gradually turns into wine red. After continuing to stir for 8-10 minutes, turn off the heat source and continue stirring to cool to room temperature, and put it in a 4°C refrigerator for later use.
[0036] (3) Add 9 μL of 100 μM thiol-labeled signal probe to 300 μL of the gold nanoparticle solution in step 2 above, and stir overnight at 4°C for 12 hours on a magnetic stirrer. The signal probe sequence is: 5'-SH-(CH 2 ) 6 -TTTTTTCAGAACTCACCTGTTAGTTTTTT-biotin-3'.
[0037] (4) Add 0.5M NaCl to the gold nanopa...
Embodiment 3
[0039] Embodiment three, prepare the sample to be tested
[0040] 1. Extraction of genomic DNA used as PCR template
[0041] Because many Salmonella infections are foodborne, food or feces can usually be used as a source of detection.
[0042] Food testing: operate under sterile conditions, put the food to be tested into a homogenizer and stir into a homogenate, take 25g (or mL) of the homogenate of the sample to be tested and dilute it with 225mL buffered peptone water (BP) to 1 : 10 homogeneous slurry. Take 1 mL of the homogeneous liquid in a centrifuge tube, centrifuge at 12,000 r / min for 5 min, wash with sterilized saline twice, and finally suspend with 0.25 mL of distilled water, incubate in a 100°C water bath for 15 minutes, and then immediately place on ice. After centrifuging at 12000 r / min at 4°C for 10 minutes, the supernatant was transferred to a new tube, and the supernatant contained genomic DNA that could be directly used as a PCR template.
[0043] Stool dete...
PUM
| Property | Measurement | Unit |
|---|---|---|
| particle size | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com