Genetic recombinant saccharomyces cerevisiae capable of degrading and utilizing kitchen wastes
A technology for kitchen waste and Saccharomyces cerevisiae, which is applied in the fields of genetic engineering and fermentation engineering, can solve the problems of polluted environment, lack of degradation, rotten and smelly kitchen waste, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0033] Example 1 α-amylase gene amy , glucoamylase gene ga and acid protease gene ap clone
[0034] Refer to Aspergillus oryzae published on GenBank ( Aspergillus oryzae ) α-amylase gene amy (accession number XM_001821384), Aspergillus niger ( Aspergillus niger ) glucoamylase gene ga (accession number XM_001390493.1) and acid protease gene ap (Accession No. XM_001401056.2), use Oligo 6 primer design software to design primers, and add appropriate restriction sites at the same time:
[0035] amy Gene amplification primers:
[0036]
[0037] The total RNA of Aspergillus oryzae CICC 40344 was extracted, and the reverse transcription PCR amplification reaction was carried out. amy The PCR amplified product of the gene was connected to the pGEM-T Easy vector (purchased from Promega Company) and verified by sequencing.
[0038] in amy The PCR reaction conditions of the gene are:
[0039]
[0040] The total RNA of Aspergillus niger CICC 40179 was extracted,...
Embodiment 2
[0046] Example 2 Three kinds of enzyme gene construction co-expression recombinant vector
[0047] The construction process of the three enzyme gene co-expression recombinant plasmids is as follows: figure 1 shown.
[0048] Obtained by embodiment 1 amy , ga with ap Restriction enzymes for coding sequences Bam H I and Speech I were excised from the pGEM-T Easy vector with double enzymes, respectively, and connected to the vector pScIKP that had been digested with the same double enzymes, to obtain recombinant plasmids pScIKP-amy, pScIKP-ga and pScIKP-ap.
[0049] use Nhe I and Xba I double digested pScIKP-ga, obtained containing PGK promoter and terminator ga Gene expression cassette fragments. use Nhe I single digested pScIKP-amy to linearize it. The two were ligated with T4 DNA ligase (using Nhe I and Xba I is the principle of the same tail enzyme), and the recombinant plasmid pScIKP-amy-ga was obtained. Using the same principle, use Nhe I an...
Embodiment 3
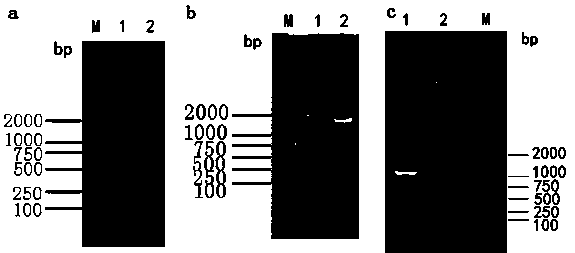
[0050] Example 3 Screening and Validation of Recombinant Yeast Transformants
[0051] Before the electrotransformation of Saccharomyces cerevisiae, the sensitivity of Saccharomyces cerevisiae AS2.489 was tested for the sensitivity of the resistance screening marker G418, and it was found that the yeast had been inhibited and could not grow on the YPD plate with a G418 concentration of 150 μg / ml. When the child can be screened with a concentration of G418 exceeding 150 μg / ml.
[0052] The three gene co-expression recombinant plasmid pScIKP-amy-ga-ap that embodiment 2 obtains uses restriction endonuclease Apa After I was linearized, it was transformed into Saccharomyces cerevisiae AS2.489 by electroporation, and cultured on a YPD agar plate with a G418 concentration of 200 μg / ml for 3 to 4 days, and the colony that could grow normally was picked as the transformation. Transformants of the above recombinant plasmids. Colony PCR was carried out with primers specific to the thr...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com