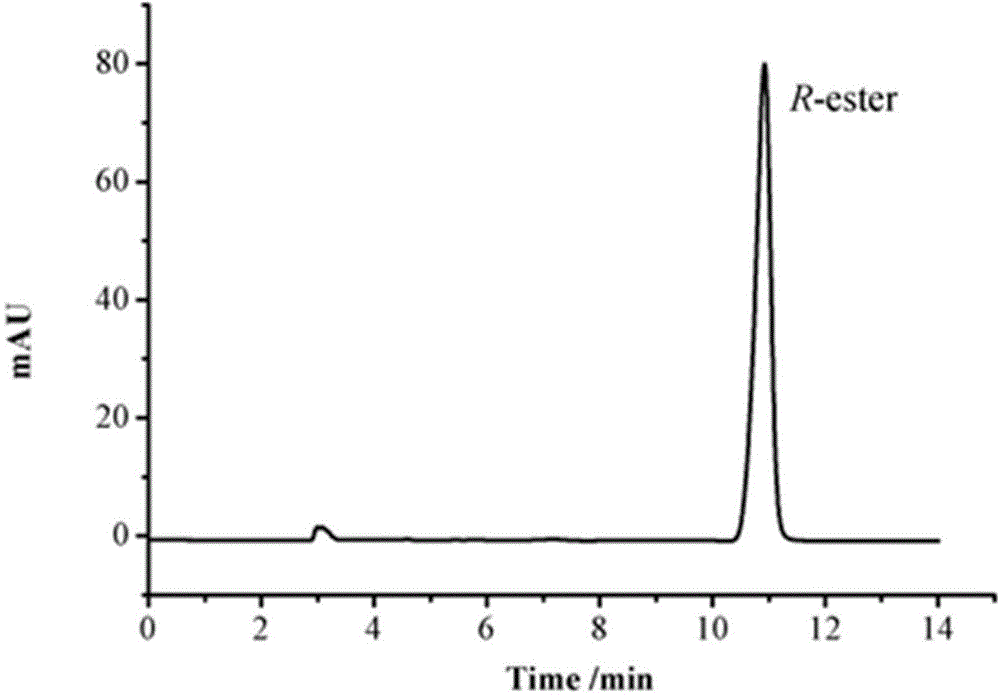
Method for preparing R-o-chloromandelic acid methyl ester through biocatalysis dynamic kinetic resolution
A technology of methyl mandelate and dynamic kinetics, applied in biochemical equipment and methods, methods based on microorganisms, microorganisms, etc., can solve problems such as lack of, and inability to achieve ideal yields, and achieve simple operation, low production cost, The effect of high catalytic efficiency
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
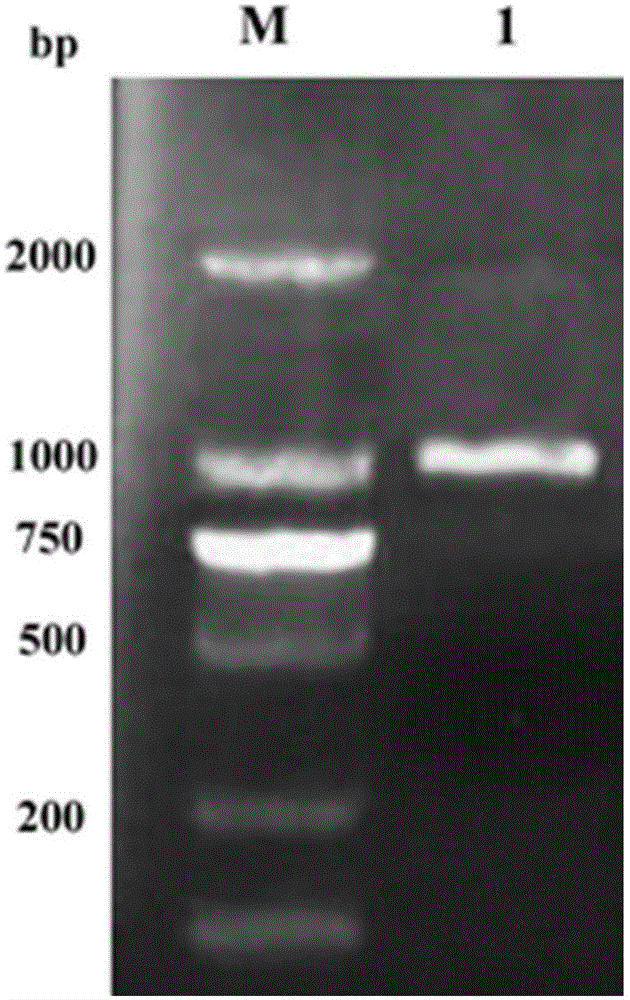
[0035] Embodiment 1, to the cloning of mandelate racemase gene
[0036] According to the mandelic acid racemase gene sequence (mdla) of Pseudomonas putida in NCBI, the upstream and downstream amplification primers and enzyme cutting sites EcoR I / Xho I were designed (the parts in italics are the enzyme cutting sites and corresponding protection base). Primers were designed as follows:
[0037] Primer 1: GGAATTC ATGAGTGAAGTACTGATTACCG
[0038] Primer 2: CCGCTCGAGTTTACACCAGATATTTCCCGATTT
[0039] PCR amplification was performed using the genomic DNA of Pseudomonas putida (ATCC12633, purchased from the American Type Culture Collection) as a template. The PCR system is 10×DNA polymerase buffer5μL, MgCl 2 (25mmol / L) 4μL, dNTPs (10mmol / L) 1μL, primer 1 (10μmol / L) 1μL, primer 2 (10μmol / L) 1μ, genome template: about 10ng, Taq DNA polymerase (5U / μL) 0.5μL, ddH 2 O (i.e. redistilled water) to a total volume of 50 μl.
[0040] The PCR amplification steps are: (1) pre-denaturation a...
Embodiment 2
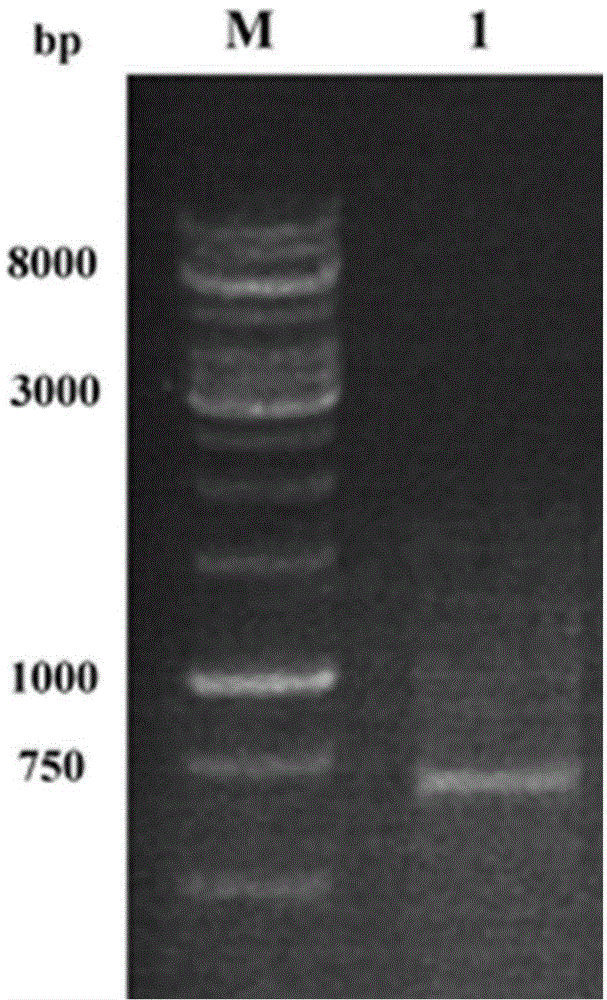
[0041] Embodiment 2, preparation of recombinant plasmid pET30a-MR
[0042] The mandelic acid racemase gene target band recovered in Example 1 was double-digested with restriction endonuclease EcoR I / Xho I for 12 h at 37°C, purified by agarose gel electrophoresis, and purified using agarose gel DNA The recovery kit recovers the target band. Under the action of T4 DNA ligase, the target fragment was ligated with the plasmid pET30a that had also been digested with EcoR I / Xho I, and the recombinant plasmid pET30a-MR was obtained overnight at 4°C.
Embodiment 3
[0043] Embodiment 3, the construction of recombinant MR mutant strain
[0044] Using the recombinant MR obtained in Example 2 as a template, use the QuickChange method to perform site-directed mutagenesis at the 22 or 26 position. The PCR amplification steps are: (1) pre-denaturation at 98°C for 1 min, (2) at 98°C Under the condition of ℃, denature for 30s, (3) anneal at 56℃ for 90s, (4) extend at 72℃ for 7min; repeat steps (2)-(4) 20 times, (5) extend at 72℃ for 5min, cool to 4℃. The obtained PCR product was cleaned and digested with Dpn I to eliminate the methylated template, and then retransformed into E.coli BL21 (DE3) competent cells, and spread onto LB plates containing kanamycin. Cultured upside down at 37°C overnight, single clones were picked and sequenced for verification, and recombinant MR mutants were obtained.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com