Primer for rapidly detecting HLA-B*1502 allele, kit and detection method
A technology of HLA-B and reagent kit, which is applied in the field of genetic engineering, can solve the problems of prone to false positives, failure to meet high specificity detection, and few genotypes, so as to save operation time and workload and prevent false positives The effect of producing and experimenting with low cost
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0024] Example 1: Design of primers for rapid detection of HLA-B*1502 gene
[0025] The data of all HLA alleles required for PCR primer design comes from IMGT / HLADatabase (Release3.15.0, 2014-01-17), the specific website is: http: / / www.ebi.nc.uk / ipd / imgt / hla / . The primers were designed manually, and the designed primers were compared in the IMGT database to confirm that the primers could specifically bind to the HLA-B*1502 allele. In the primer design process, the key is to make the designed primers able to specifically amplify the HLA-B*1502 gene in a specific PCR buffer system environment, that is, the primer is a "sequence-specific SSP".
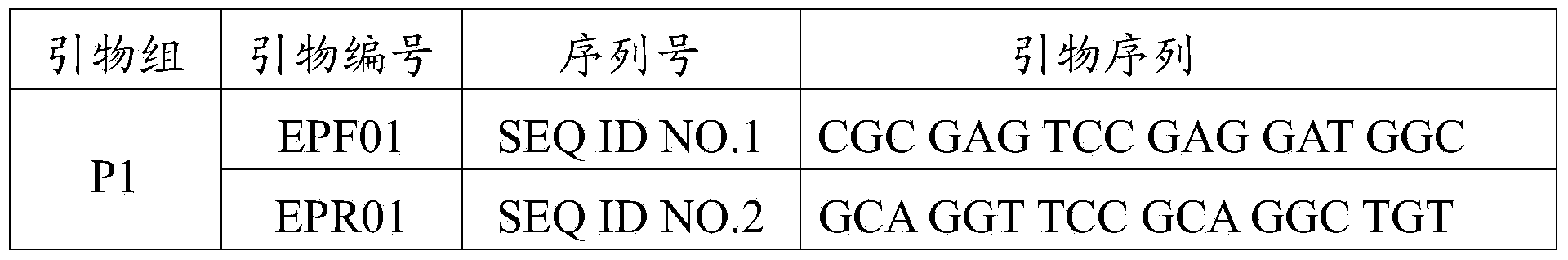
[0026] In order to use specific PCR-SSP to screen the HLA-B*1502 gene, specific primers EPF01 (SEQ ID NO.1) and primer EPR01 (SEQ ID NO. ID NO.2), primer EPF02 (SEQ ID NO.3) and primer EPR02 (SEQ ID NO.4), the four primers were combined into two groups, P1 and P2, to prevent the missed detection of the HLA-B*1502 gene, The P1 group is...
Embodiment 2
[0034] Example 2: Preparation of a kit for rapid detection of HLA-B*1502 gene
[0035]1. Synthesis of primers
[0036] The primers were synthesized by Shanghai Yingjun Biotechnology Co., Ltd., and the sequences of the primers are respectively shown in SEQ ID NO.1-6, and the specific sequences are shown in Table 1.
[0037] 2. Preparation of PCR reaction mixture
[0038] Prepare primer sets P1 and P2 respectively as shown in Table 1, and mix each primer shown in P1 set and P2 set with internal control primers, dNTPs, dye cresyl red, and PCR buffer respectively. The concentrations of detection primers SEQ ID NO.1-4 are all 0.5 μM, the concentrations of internal reference primers SEQ ID NO.5-6 are respectively 0.2 μM, the concentration of dye cresyl red is 0.01%, and the concentration of dNTPs is 0.2 mM. The PCR buffer solution is: Tris-HCL at a concentration of 10 mM, potassium chloride at a concentration of 50 mM, magnesium chloride at a concentration of 1.5 mM, and gelatin a...
Embodiment 3
[0039] Example 3: Typing detection of HLA-B*1502 gene in samples
[0040] Eight DNA samples with known HLA-B gene results were selected. Add to two PCR tubes containing the P1 and P2 primer sets respectively, and add Taq enzyme to each tube at the same time. After adding the sample, mix the reaction mixture evenly, centrifuge briefly, and carry out the PCR reaction.
[0041] The conditions of the PCR reaction were: 94°C for 5 minutes; followed by 30 cycles of 94°C for 1 minute, 65°C for 2 minutes, and 72°C for 1 minute; finally, 72°C for 10 minutes, then cooled to 15°C for gel electrophoresis detection.
[0042] Use 0.5×TBE buffer to prepare 2% agarose gel. Take 3ul of PCR products and directly spot on the gel well, run electrophoresis at 150V for 40 minutes, and then take pictures and record under ultraviolet light. For specific gel electrophoresis pictures, please refer to the attached figure 1 .
[0043] Interpretation of the results: When the internal control bands app...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com