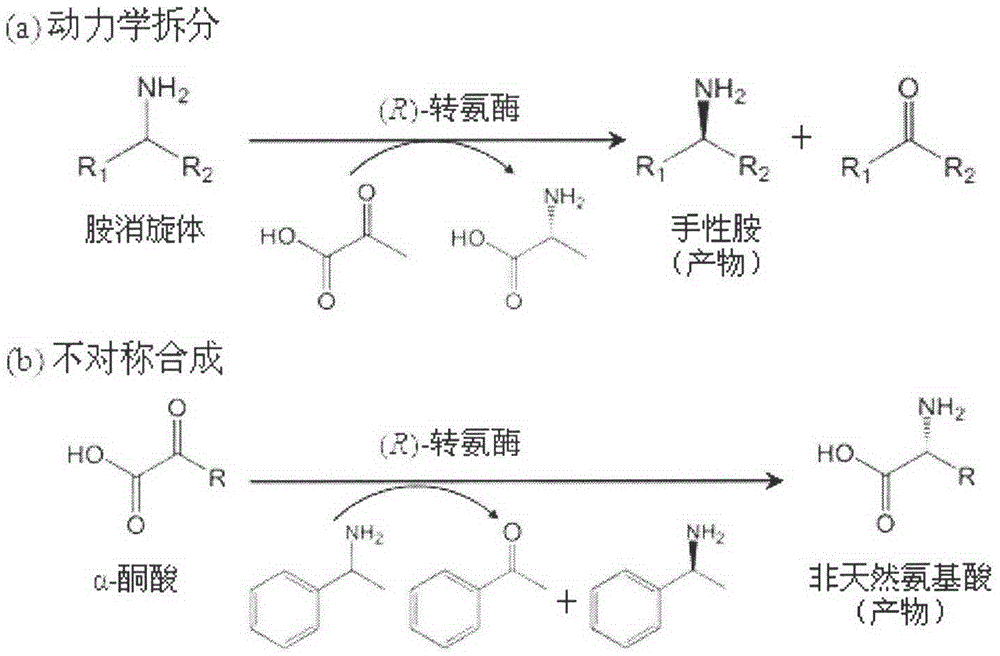
New (R)-transaminase from Trichoderma reesei and application thereof
A transaminase and amino acid technology, which is applied in the direction of transferase, enzyme, and the introduction of foreign genetic material using carriers, etc., can solve the problems of transaminase and other problems, and achieve good temperature stability and broad substrate spectrum.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0028] Embodiment 1 The preparation method of new (R)-transaminase HTR
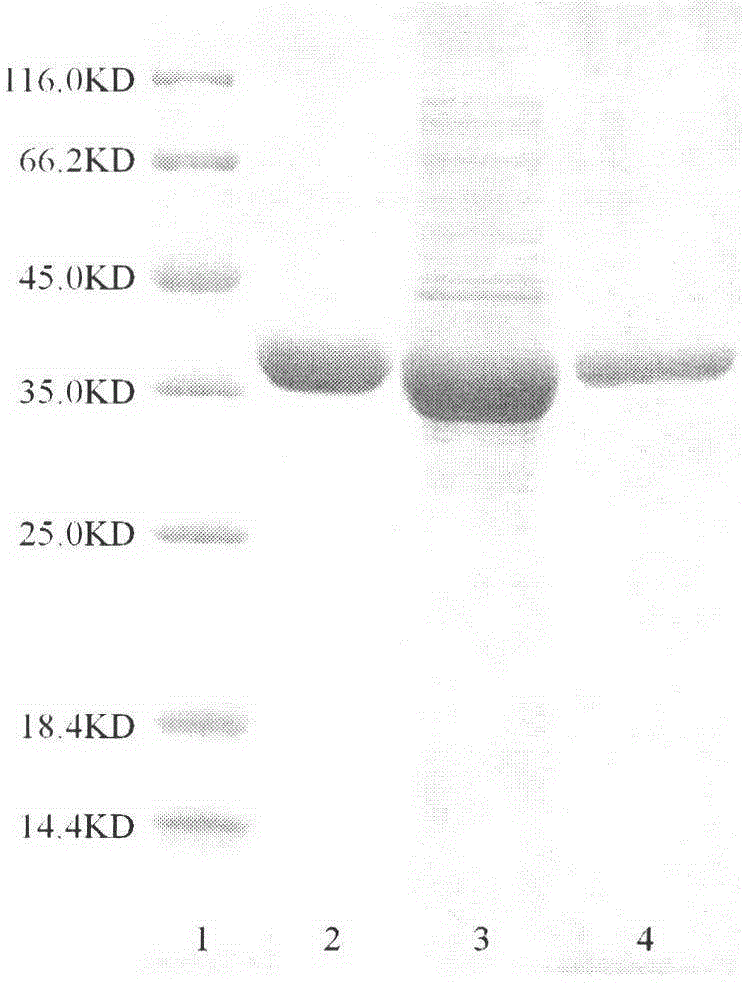
[0029] (1) Construction of engineering bacteria: The coding gene (SEQ NO.1) of the new (R)-transaminase HTR was obtained by total gene synthesis (codon optimization was carried out according to the codon preference of Escherichia coli, and the N-terminal was added with 6×His-tag in order to purify the target protein (HTR) with a nickel affinity column), and connect to the pET32a expression vector Nde I / BamH I site. The recombinant plasmid is transferred into the host bacterial cell (preferably Escherichia coli BL21 (DE3)), and the corresponding engineering strain is obtained.
[0030] (2) Expression and purification: LB medium was used to culture in a shaker at 37°C until logarithmic phase, and after cooling to 25°C, IPTG with a final concentration of 0.1 mM was added to induce expression in a shaker at 25°C for 12 hours. Collect the bacteria by centrifugation at 4°C and 6,000 rpm, wash twice with sodium p...
Embodiment 2
[0032] Example 2 Enzymatic properties of new (R)-transaminase HTR
[0033] The optimum pH value, optimum reaction temperature and temperature stability of the new (R)-transaminase HTR were examined.
[0034] Simple reaction formula:
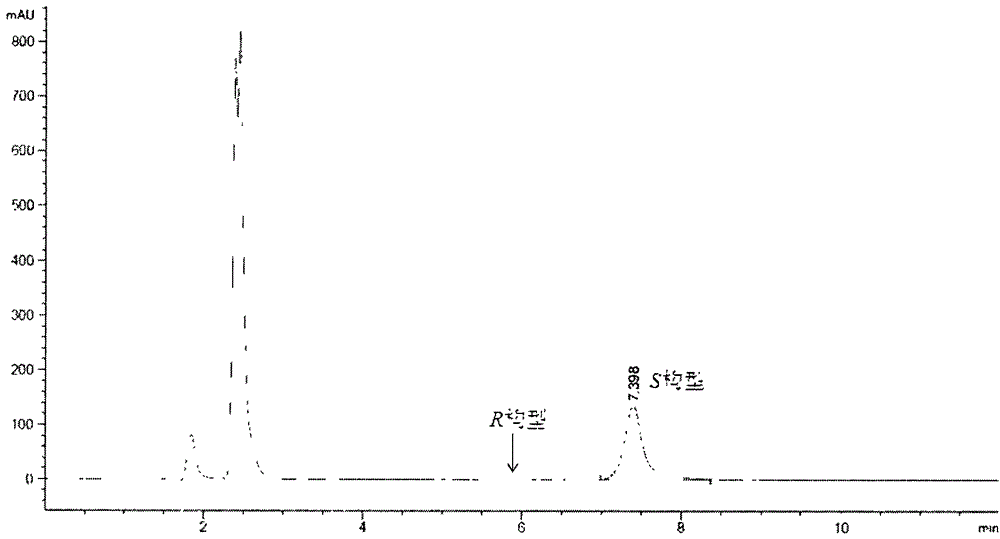
[0035] Activity assay reaction system: 1 ml of pH buffer, PLP (20 μM), (R)-1-phenylethylamine (10 mM), pyruvic acid (10 mM). The reaction was initiated by adding an appropriate amount of neo(R)-transaminase HTR, and terminated with perchloric acid after half an hour of reaction. After the reaction was terminated, the amount of acetophenone generated by the reaction was determined by HPLC to determine the size of the enzyme activity.
[0036] Determination of the optimum reaction pH value: adopt the bioassay reaction system, the reaction temperature is 30°C, measure at a series of different pH values (pH=5, 6, 7, 7.4, 7.8, 8, 8.4, 8.8, 9, 9.4, 9.6 , 10) The enzymatic activity of the new (R)-transaminase HTR. The buffers used in different p...
Embodiment 3
[0040] Example 3 The substrate spectrum and application of the new (R)-transaminase HTR
[0041] (1) Amino donor substrate spectrum of the new (R)-transaminase HTR.
[0042] Simple reaction formula:
[0043] Reaction conditions: 1ml of reaction system. Add pyridoxal phosphate (20μM), amino donor substrate (10mM, racemate 20mM) and pyruvate (10mM) into sodium phosphate buffer (100mM, pH7.4), mix well and add appropriate amount of HTR enzyme solution Start the reaction, react in a 30°C water bath for 30 minutes, and then terminate the reaction with 375ul of 16% perchloric acid.
[0044] Calculation of relative activity: Calculate the relative activity of each substrate by detecting the decrease of pyruvate in the reaction system. The liquid phase detection conditions of pyruvate are: Aminex HPX-87H column (Bio-Rad, USA), column temperature 45 ° C, with 5 mM H 2 SO 4 As the mobile phase, the flow rate is 0.6ml / min, and the ultraviolet absorption value at 205nm is detected....
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com