A method for producing 1,3-propanediol by fermentation of glycerol with mixed bacteria
A technology of mixed bacterial fermentation and propylene glycol, applied in the field of bioengineering, can solve the problems of not completely preventing production, inhibiting cell growth, increasing lactic acid, etc., and achieves the effects of simple operation, good substrate tolerance and high production intensity.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0057] Example 1 Mixed bacteria screening, strain isolation and identification
[0058] 1. Screening of mixed bacteria
[0059] Randomly collect 40 samples of barnacles, seaweed, sponges and sea mud in the sea area near Dalian, in a sterile ultra-clean bench, get 2g of each sample (barnacles, seaweeds, sponges need to be ground into a slurry with a sterile mortar, sea mud without pretreatment), were added to 250mL Erlenmeyer flasks containing 50mL enriched medium, and the bottle mouths were sealed with gauze and kraft paper. Cultivate on a shaker at 37°C and 200r / min for 24 hours to obtain the primary culture; then inoculate 1 mL of the primary culture into another Erlenmeyer flask containing the seed medium, and cultivate under the same conditions for 24 hours to obtain the secondary culture; The third enrichment was carried out under the same conditions.
[0060] The enriched bacterial liquid of each sample obtained above was screened by shaking flask fermentation, and t...
Embodiment 2
[0066] The batch fermentation of embodiment 2 mixed bacteria NHN8
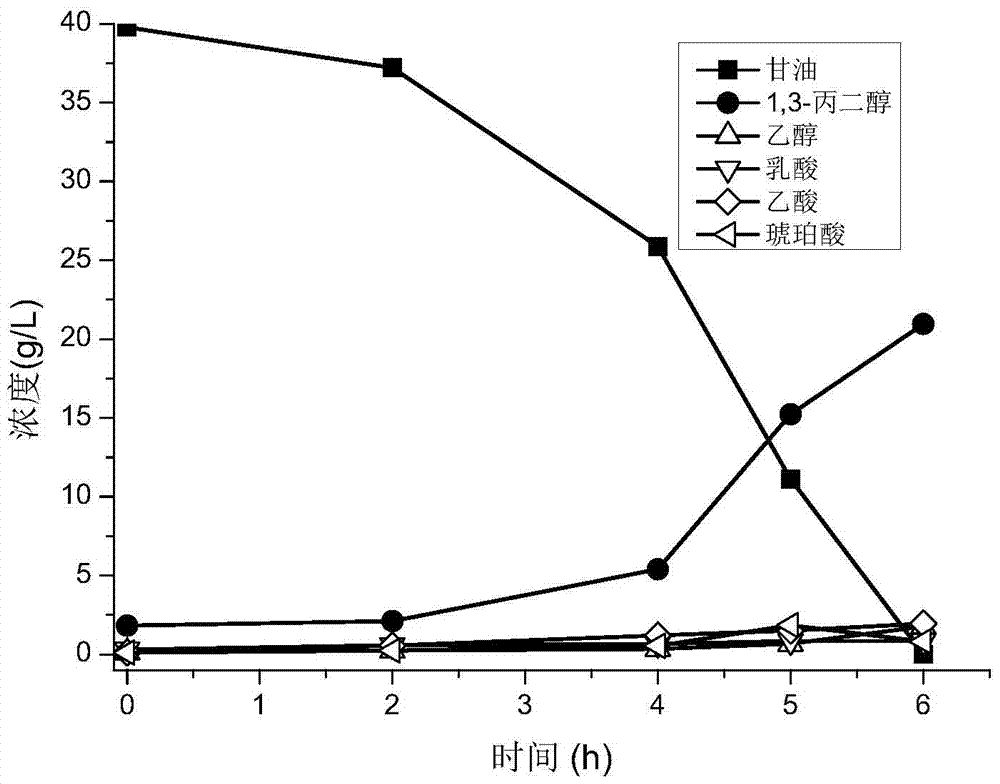
[0067] The mixed bacteria NHN8 obtained in Example 1 (the thalline number ratio of three kinds of bacteria contained therein is W3:Y1:Y5=208:17:82) is inserted in the 250mL Erlenmeyer flask that 100mL seed culture medium is housed, bottle The mouth was sealed with gauze and kraft paper, and the seeds were cultured in a shaker at 37°C and 200r / min for 12 hours to obtain the seed culture solution; then the seed culture solution was inoculated into a 5L fermenter with 2L fermentation medium at a volume ratio of 10% , the fermentation medium is steam sterilized in advance and then cooled. Fermentation was carried out at 37°C, 250r / min, nitrogen flux 0.2vvm, and pH 7.0. During the fermentation process, take the fermentation broth every 1 or 2 hours, measure the glycerol concentration with sodium periodate oxidation method, measure the OD of the bacteria with spectrophotometry, and centrifuge the fermentation broth...
Embodiment 3
[0069] The batch fermentation of embodiment 3 single bacterium W3
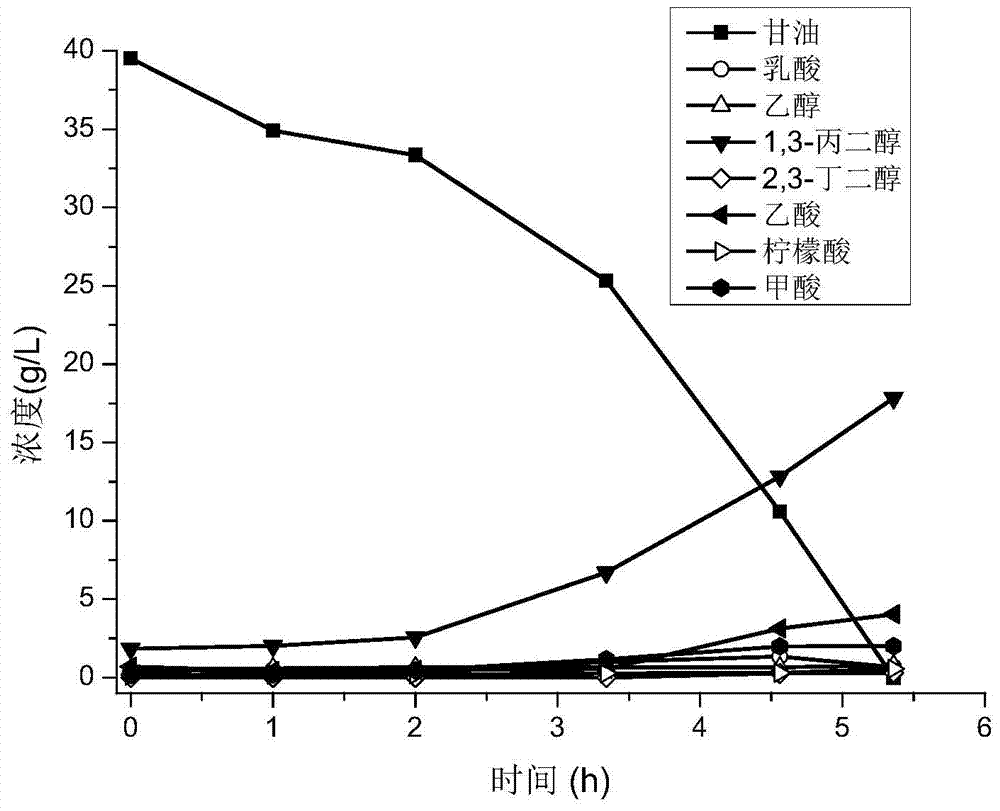
[0070] According to the method described in Example 2, the single bacterium W3 obtained in Example 1 was inserted into the seed medium and cultivated for 12 hours to obtain the seed culture solution, and then the seed culture solution was inoculated into a 2L fermentation tank according to the volume ratio of 10%. The medium was fermented in a 5L fermenter at 37°C, 250r / min, nitrogen flux 0.2vvm, and 5M NaOH to maintain the pH at 7.0. During the fermentation process, the fermentation broth was taken every 1 or 2 hours to measure the glycerol concentration and cell OD, and the fermentation broth was centrifuged at 10000r / min for 10 minutes, and the supernatant was taken for high-performance liquid chromatography analysis.
[0071] Fermentation results such as image 3 As shown, the glycerin of 40g / L was consumed by strain W3 in 5.3 hours of fermentation, the PDO concentration reached 17.84g / L, the production i...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com