Xylanase, gene coding same and application of xylanase to waste paper deinking
A xylanase, gene technology, used in applications, glycosylase, processing waste paper, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0038] Embodiment 1: the acquisition of xylanase gene, the construction of recombinant plasmid and recombinant engineered bacteria:
[0039] Obtaining the xylanase gene:
[0040] Analyze, screen, discover using the National Center for Biotechnology Information (NCBI) database Planomicrobium glaciei There is a sequence in CHR43 with BacillusThe sequence of xylanase (Genbank number: AAB70918) in sp.StrainNG-27 has high homology, and it is likely to be the gene sequence encoding xylanase. The sequence is screened and optimized (optimization includes mutation and insertion of some sites), and the nucleotide sequence with SEQ ID NO.2 is obtained. The xylanase gene was artificially synthesized by GenScript according to the nucleotide sequence of SEQ ID NO.2, and BamHI and XhoI restriction sites were added at both ends of the sequence, and then connected to the BamHI and XhoI restriction sites on the pETDuet-1 plasmid vector Between the sites, the recombinant plasmid pETDuet-1-xy...
Embodiment 2
[0044] Expression and purification of embodiment 2 xylanase xyn2
[0045] Express:
[0046] Recombinant bacteria were first inoculated in 10 ml LB liquid medium containing 100 μg / ml ampicillin, and cultured overnight at 37° C. and 220 rpm with shaking. Then transfer to 500ml LB liquid medium containing 100μg / ml ampicillin at 1% amount, shake culture at 37°C and 220rpm until OD 600 When it reaches about 0.6, add IPTG to a final concentration of 1 mM, shake and culture at 37° C. and 220 rpm for 6 hours. Bacteria were collected by centrifugation.
[0047] purification:
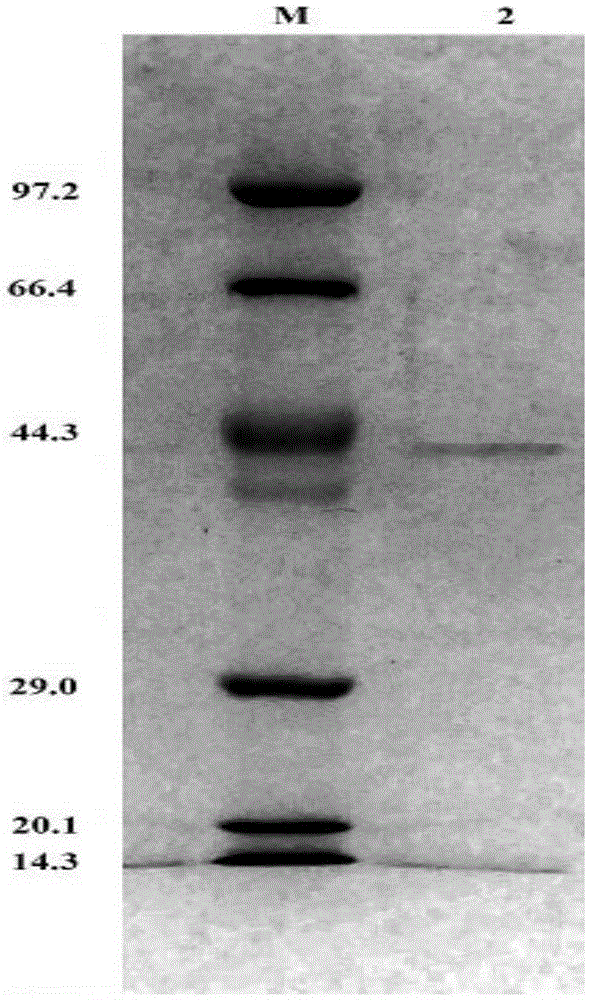
[0048] Add an appropriate amount of pH7.0PBSbuffer (137mmol / LNaCl, 2.7mmol / LKCl, 4.3mmol / LNa 2 HPO 4 , 1.4mmol / LKH 2 PO 4 , adjust the pH value of the solution to 7.0 with HCl), resuspend the recombinant bacteria by ultrasonication, centrifuge at high speed, and collect the supernatant. Incubate the supernatant at 65°C for half an hour to precipitate a large number of non-thermostable host bacterial prote...
Embodiment 3
[0049] The enzymatic property determination of embodiment 3 xylanase
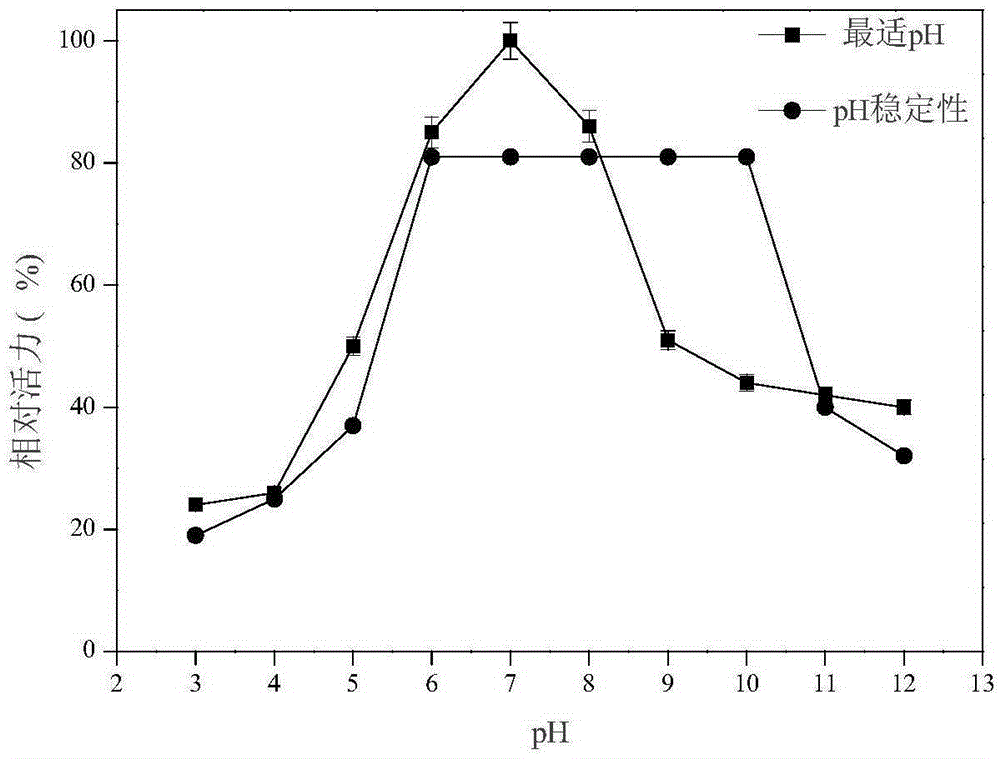
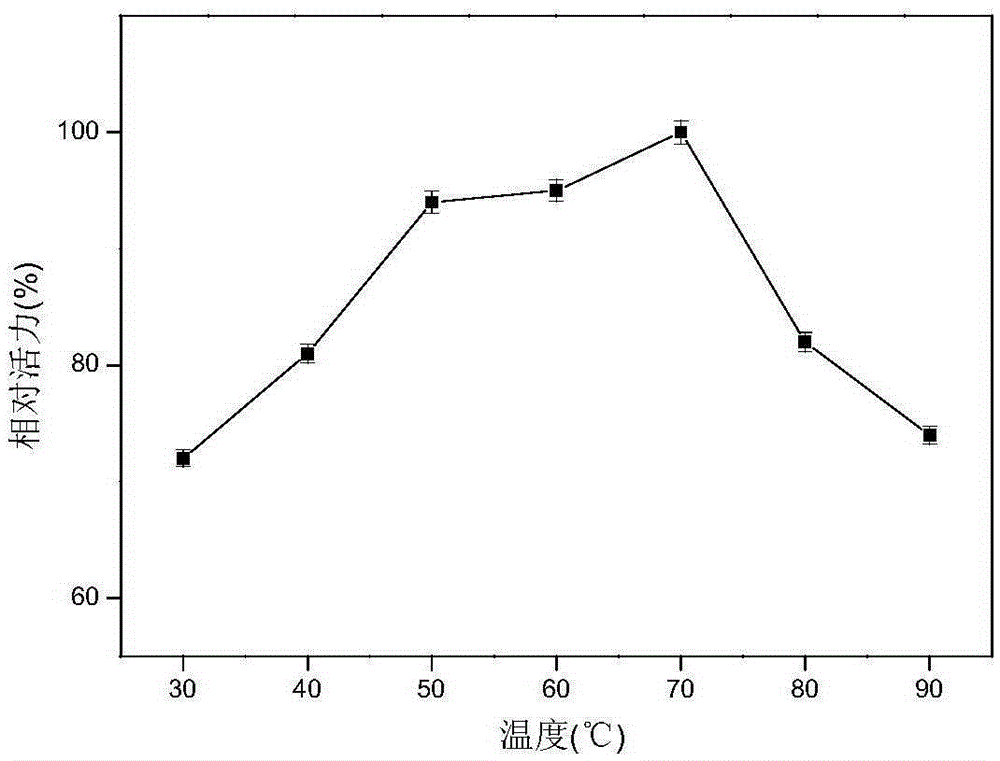
[0050] Use the DNS method to measure xylanase activity: add 200 μL of enzyme to the 2 mL reaction system, the substrate concentration is 0.9%, react at a temperature of 50 ° C and pH 4.8 for 10 min, add 2 mL of DNS to terminate the reaction, and bathe in boiling water for 10 min. After cooling to room temperature, add water to 15mL, and then detect its absorbance at a wavelength of 540nm, compare with the standard curve to obtain the reducing sugar content, and the enzyme activity unit (U) is defined as the amount of enzyme that releases 1mM reducing sugar per minute. According to this method, the activity of xylanase in the pH range of 3.0 to 12.0 was determined. The result is as figure 2 As shown, it can be seen from the figure that the optimal pH of xylanase is 7.0, and maintains a relatively high activity in the range of pH 6.0-10.0. The activity of xylanase in the range of 30°C to 90°C was also dete...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com