Novel deep-sea low-temperature salt-tolerant esterase and application
An esterase, amino acid technology, applied in the field of genetic engineering, to achieve the effect of excellent enzymatic properties
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0021] The acquisition of embodiment 1 esterase gene e10
[0022] Bacteria Croceicoccus marinus E4A9 based on deep-sea sediments T The whole genome, open reading frame prediction and gene annotation results are used to screen lipohydrolase-related genes. The homology between the sequence and the known esterase gene sequence in the database was compared by Blastx (http: / / blast.ncbi.nlm.nih.gov / ). The e10 gene was obtained through database comparison analysis, with a size of 618bp and a base composition of 106A (17.15%), 97T (15.70%), 203C (32.85%) and 212G (34.30%), and its nucleotide sequence is as shown in SEQIDNo: 1 shown. The size of the encoded protein is 205 amino acid residues, and its amino acid sequence is shown in SEQ ID No.2. A homology search was performed on the gene sequence in GenBank, and the highest similarity was the esterase in the same genus Croceicoccusnaphthovorans, with a similarity of 64%. Its registration number in the GenBank database was WP_0478202...
Embodiment 2
[0025] The construction of the recombinant expression plasmid of embodiment 2 esterase gene e10 and recombinant bacterial strain
[0026] The esterase gene e10 obtained in the present invention is cloned into an expression vector to construct a recombinant expression strain. Based on the open reading frame sequence of the esterase gene obtained by ORF analysis of NCBIORFFinder, the upstream primer E10F (5'-TCGC GGATCC GTGGCGGACGGCGAGGC-3', BamHI) and downstream primer E10R (5'-TCCG CTCGAG CTAGAGGTCGTCGATCCTGTC-3', XhoI), PCR amplification confirmed the full-length sequence of the gene. The expression plasmid was constructed by enzyme digestion cloning, that is, the PCR product was double-digested with BamHI and XhoI, and the purified fragment was ligated with the plasmid pSMT3 that had been double-digested with BamHI and XhoI. 2 The transformation method was transformed into E.coliDH5α, and positive clones were screened for kanamycin resistance. A plasmid extraction kit (...
Embodiment 3
[0027] Example 3 Utilize recombinant expression strain to express recombinant esterase gene e10
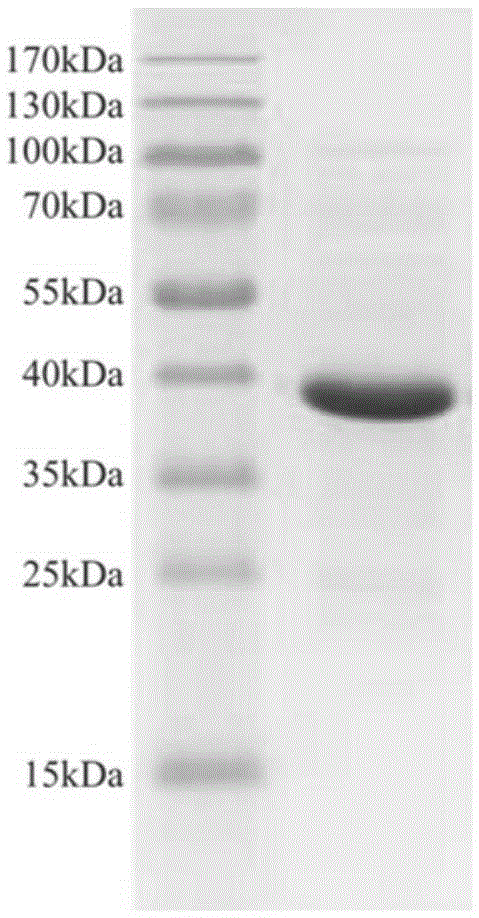
[0028] Transfer 3ml of the constructed recombinant expression strain to 100ml of LB liquid medium containing 20μg / ml kanamycin and 34μg / ml chloramphenicol, and shake at 37°C until OD 600 When it reaches 0.6, add IPTG with a final concentration of 0.5mM to induce expression, transfer to 25°C and shake at 150r / min for 8h. The bacterial cells were collected by low-temperature centrifugation, resuspended in NTA-10 solution (500 mM sodium chloride, 10 mM imidazole, 20 mM Tris hydrochloric acid, pH 8.0), and subjected to sonication on ice. Centrifuge at low temperature to collect supernatant, use NTA-Ni 2+ The expressed protein was purified by affinity column chromatography. The expressed recombinant protein contains N-terminal 6×Histag, which can be affinity-adsorbed to the layer suction column, and is eluted with different concentrations of imidazole solution gradient, and the eluat...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Absorbance | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com