Endochitinase and coding gene and application thereof in production of chitobiose
A technology of chitinase and coding gene, which is applied in the field of bioengineering, can solve the problems of chitinase research starting late, not realizing industrialization, and low yield of chitobiose, so as to achieve less energy consumption and promote development , the effect of high conversion rate
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0053] Embodiment 1 Discovery of chitinase gene
[0054] 1. Design of degenerate primers for PCR amplification of the conserved region of chitinase genome
[0055] Using Paenibacillus barengoltzii CAU904 as the genome template, according to the chitinase amino acid sequence reported in the GenBank database, the online software BlockMaker (http: / / blocks.fhcrc.org / blocks / blockmkr / make_blocks. html) to search for conserved regions. According to the conserved amino acid sequence of chitinase, the online design software CODHOP (http: / / blocks.fhcrc.org / codehop.html) was used to select the amino acid sequence of chitinase from Paenibacillus GH18 family reported in GenBank. For homologous alignment, select the sequence conservation regions THINYAFA and DLDWEYPV to design degenerate primers Chi2DF and Chi2DR in Table 2.
[0056] PCR reaction conditions: pre-denaturation at 94°C for 5 minutes; 20 cycles of denaturation at 94°C for 30s, annealing at 62-52°C for 30s, and extension at 72...
Embodiment 2
[0062] Embodiment 2 Expression of endochitinase gene
[0063] 1. Construction of recombinant expression vector
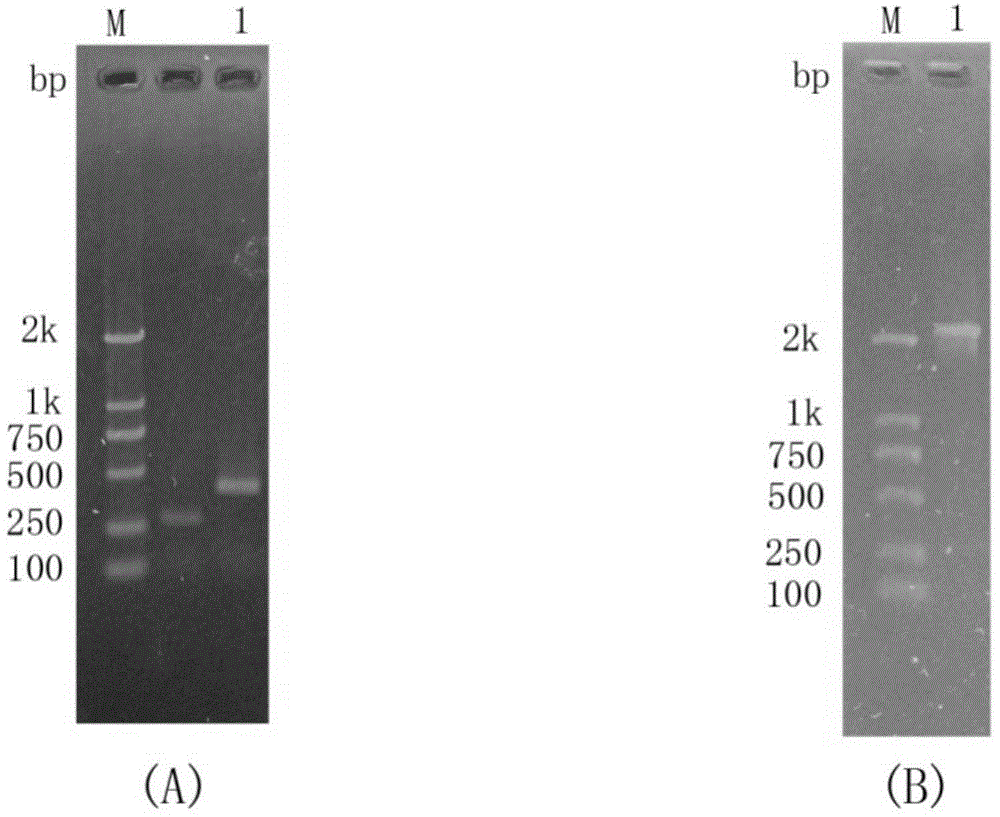
[0064] Genomic DNA of Paenibacillus barengerzia was used as a template to amplify the chitinase PbChi70 gene with the removal of the signal peptide using the upstream and downstream primers Chi2EF and Chi2ER. The agarose gel electrophoresis of the PCR amplification product is shown in figure 2 . The PCR amplification product was recovered, connected to the pMD18-T vector, and transformed into Escherichia coli DH5α by heat shock method. The positive recombinant plasmid was named pMD18-T-PbChi70 and sent for sequencing. Double-digest the plasmid pMD18-T-PbChi70, recover the target gene fragment, connect it with the expression vector pET-28a(+) that has been cut by the same double enzyme, transform the ligated product into Escherichia coli DH5α, extract the plasmid and identify the positive recombinant. The correct recombinant plasmid was named pET28a-PbChi70.
[0...
Embodiment 3
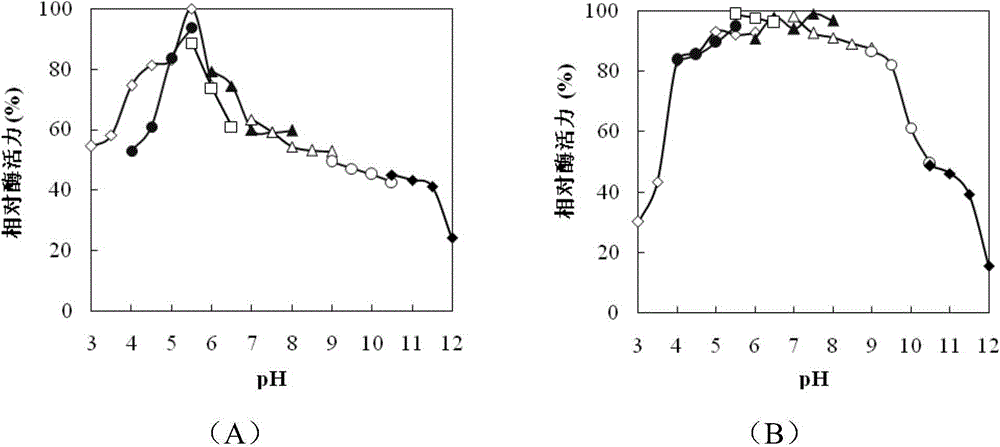
[0067] Example 3 Purification and property determination of endochitinase
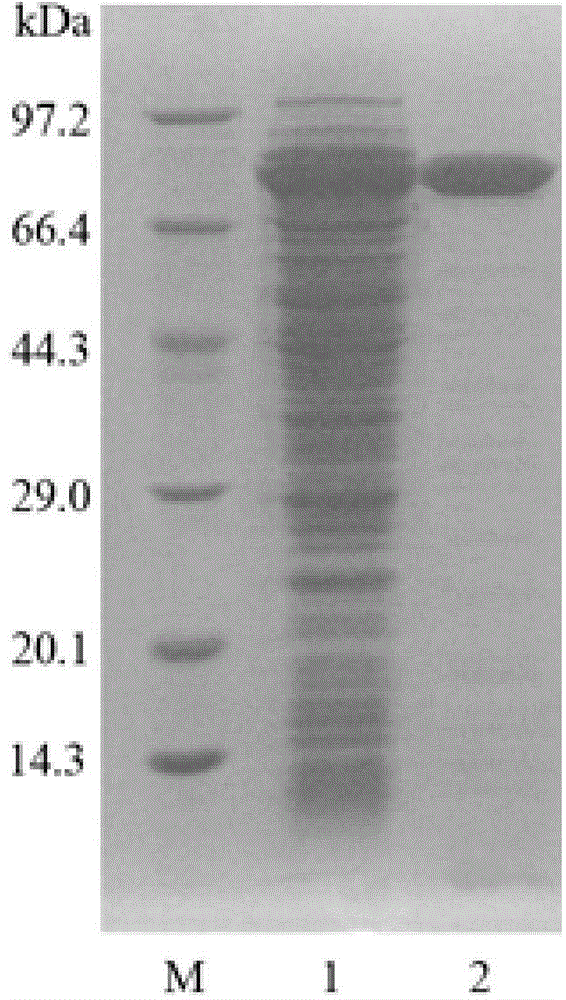
[0068] 1. Purification of Chitinase
[0069] The positive clones were picked and cultured in 50 mL LB liquid medium (50 μg / mL) at 37° C. on a shaker until logarithmic phase, and used as seed solution. Inoculate the seed liquid in 100mL LB liquid medium, use a 500mL Erlenmeyer flask containing 50μg / mL kanamycin, culture on a shaker at 37°C until the OD600 reaches 0.6-0.8, add IPTG with a final concentration of 1mM, and induce overnight.
[0070] After the culture was completed, the bacterial liquid was collected by centrifugation at 10,000 rpm for 2 minutes. The bacterial liquid was suspended in 20 mL of pH7.4 phosphate buffer, and the cells were disrupted by ultrasonication at 200 W for 3 s, intermittently for 4 s, and 90 times. After the cells are disrupted, centrifuge at 10,000 rpm for 10 minutes to collect the supernatant, namely the crude enzyme solution. The pellet was resuspended in phosphate ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com