Cyclohexanone monooxygenase and application thereof in synthesis of esomeprazole
A technology of cyclohexanone monooxygenase and cyclohexanone monooxygenase, which is applied in the field of bioengineering, can solve the problems of low yield, high cost, difficult post-treatment, etc., and achieve the effect of high yield and low price
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0047] Embodiment 1 screening cyclohexanone monooxygenase
[0048] Soil samples were collected from Shanghai Jinshan Chemical Industry Park and DNA was extracted (extraction method refers to ChromaSpinTE-1000, ClontechLaboratories, Inc., USA), partially digested with Sau3AI, and 0.5-3kb fragments were collected by electrophoresis, recovered and ligated to pWF-1 The BamHI site was used to obtain a plasmid library. Transform the library into E.coliDH5α and smear it on an LB plate containing 100 μg / mL ampicillin, select positive clones and inoculate them into a 96-deep-well plate containing 500 μg / mL LB (containing 100 μg / mL ampicillin), and incubate at 37°C for 4 hours Add 1mMIPTG for induction, and continue culturing overnight at 30°C; then take 50μL of each deep-well culture to a new 96-well plate added with 50mM sodium phosphate buffer (pH8.5) and 1mg / mL lysozyme, and incubate at 30°C for 30 minutes , down to -80°C and repeated freezing and thawing to lyse the bacteria.
[...
Embodiment 2
[0050] Expression and fermentation of embodiment 2 cyclohexanone monooxygenase
[0051] The cyclohexanone monooxygenase gene obtained in Example 1 was cloned into pET28a, and the restriction sites were NdeI-HindIII. The plasmid was transformed into E.coliBL21(DE3), positive colonies were screened on 50μg / mL kanamycin LB plate, inoculated into 200mL LB medium, and cultured at 37°C, 180-220rpm for 10-16h. The above-mentioned cultivated seeds were inoculated in a 3L upper tank fermentation medium (M9, containing glucose 4g / L, disodium hydrogen phosphate 12.8g / L, potassium dihydrogen phosphate 3g / L, Ammonium chloride 1g / L, sodium sulfate 0.5g / L, calcium chloride 0.0152g / L, magnesium chloride hexahydrate 0.41g / L), at 25~35℃, 300~800rpm, air flow 2~6L / min cultivated under conditions. After culturing for 6-10 hours, feed medium containing 60% glycerol was fed at a rate of 5-20 mL / h until the end of fermentation. Fed feed medium for several hours to OD 600 When reaching 20-40, add...
Embodiment 3
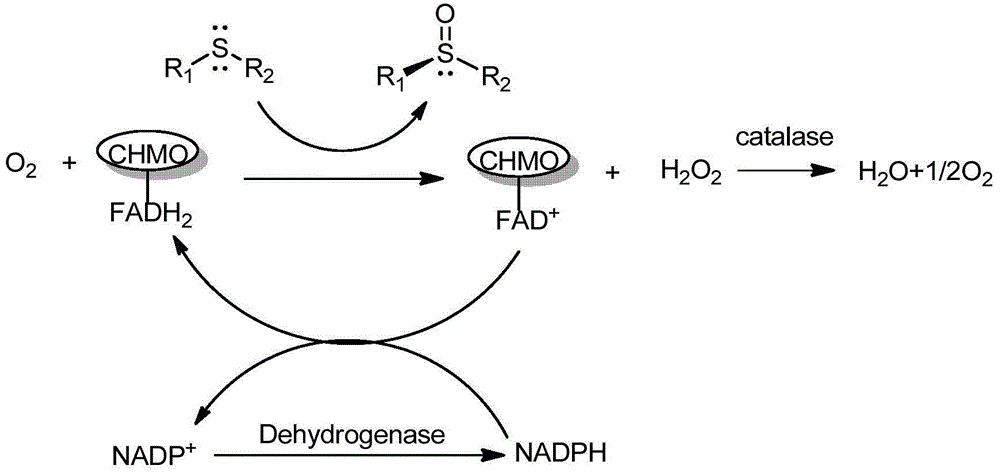
[0052] The assay method of embodiment 3 enzyme activity
[0053] The bacteria were resuspended in PBS buffer (pH9.0) at a ratio of 1:10, and after high-pressure crushing, 50 μL of crude enzyme solution was added to 1.8 mL of PBS buffer (pH9.0), and methanol containing 100 mM cyclohexanone was added to aid dissolution. , so that the final concentration of cyclohexanone is 10 mM; add NADPH 0.1 mM to start the reaction, and detect the change of ultraviolet absorption at a wavelength of 340 nm.
[0054] Calculation of enzyme activity: U=ΔA 340 ×1000 / (6220×20), that is, the specific enzyme activity per mL of lysate.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com