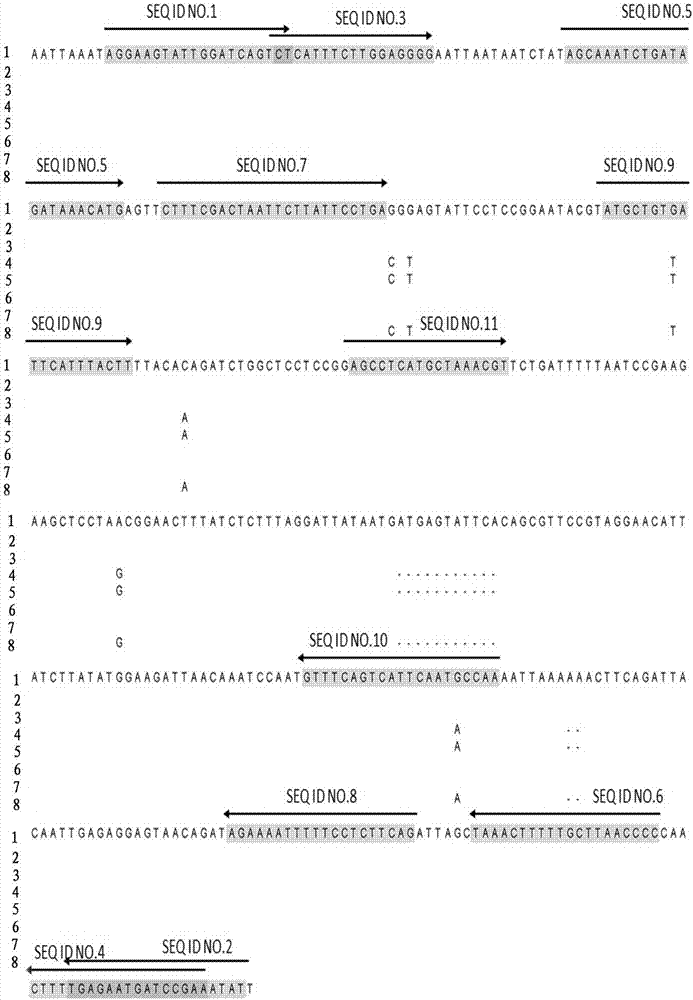
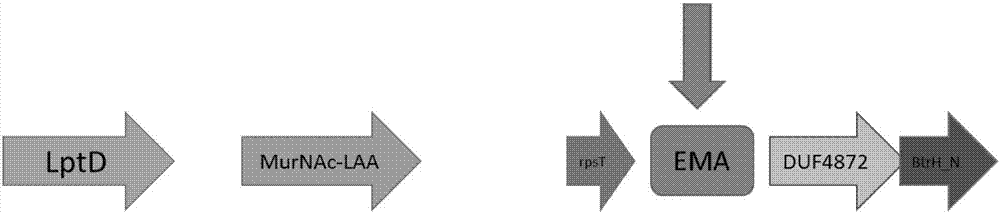
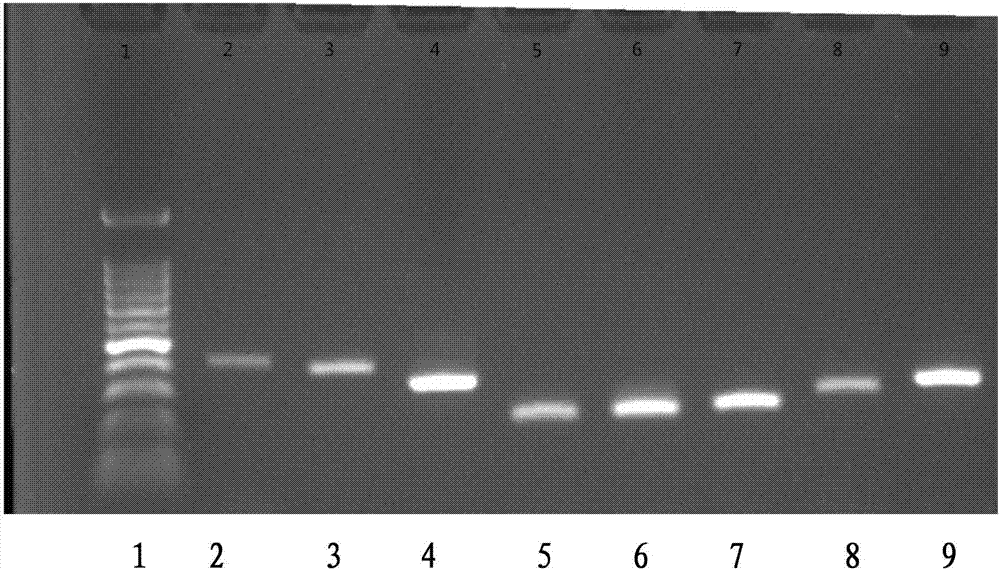
Primer combination for detecting PCR of Elizabethkingia meningoseptica
A technology of primer sequence and amplification primer, which is applied in the field of microbiology and molecular biology, and achieves the effects of wide application prospects, excellent specificity and sensitivity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0076] Example 1: Establishment of PCR amplification system
[0077] Common PCR reagents and reaction systems and conditions:
[0078] Premix Taq Mix (TaKaRa lot: RR901A); primers were designed by Primer3.0 software and synthesized by Aoke Dingsheng; agarose (Biowest lot: 11176); GoldView nucleic acid stain (Solarbio lot: G8140); 100bp DNA Marker (full Formula gold lot: BM301-02); gel imaging system Bio-red GelDoc XR; SENSQUEST PCR amplifier; Eppendorf 5415D centrifuge; DYY-8C (Beijing Liuyi Instrument Factory).
[0079] reaction system:
[0080] name Volume (uL) Final concentration 2 x Taq Mix 25 upstream primer 2 400nM downstream primer 2 400nM template DNA 2 h 2 o
19 total capacity 50uL
[0081] Reaction conditions:
[0082]
[0083] SYBR Green Real-time PCR detection reagent and reaction system: UltraSYBR Mixture (Low ROX) (Kangwei Century lot: CW2601M); primers were designed by Primer3.0 software...
Embodiment 2
[0092] Embodiment 2: General PCR and SYBR Green Real-time PCR specificity verification:
[0093] Specific detection: Simultaneous detection of Elizabeth meningitidis (ATCC 13253), Neisseria meningitidis (CMCC29356), Haemophilus influenzae (CDC control isolate M5216), Staphylococcus aureus (ATCC 25923), Streptococcus pneumoniae (ATCC) 49619), Escherichia coli (ATCC 25922), Listeria monocytogenes (SLCC 2372), Mycoplasma pneumoniae (ATCC 29342), Bordetella pertussis (ATCC 9797), Klebsiella pneumoniae (CMCC 46114) and Mycobacterium tuberculosis ( H37Rv) was detected.
[0094] A, B, C, D, E, F, G, and H primer combinations were used to amplify and detect the common pathogenic bacteria that easily cause meningitis and upper respiratory tract symptoms above to verify the specificity of the primers.
[0095] The specificity detection results show that the primers of group A can produce amplification products of non-target fragment size against Haemophilus influenzae (the target fragm...
Embodiment 3
[0101] Embodiment 3: Ordinary PCR and SYBR Green Real-time PCR sensitivity verification
[0102] The nucleic acid at a concentration of 1 ng / uL of Elizabeth meningitidis (ATCC 13253) was used as the stock solution, and the diluted samples were tested by ordinary PCR and fluorescence quantitative PCR respectively.
[0103] Quantitative detection of DNA of freshly prepared Elizabeth meningitidis strain ATCC13253 and then diluted to 10 -1 ng / μL(1ng / μL≈4.28*10 5 copy / Ul), 10 -2 ng / μL, 10 -3 ng / μL, 10 -4 ng / μL, 10 -5 ng / μL, 10 - 6 ng / μL, 10 -7 ng / μL, 10 -8 ng / μL for sensitivity verification. Figure 12 Sensitivity results of A, B, C, D, E, F, G, and H primer combinations for the detection of Elizabeth meningitidis (ATCC 13253) by the common PCR method are shown. In the electrophoresis detection of each group, the lanes are from left to right. The order of arrangement is: 100bp DNA molecular weight marker, 10 -1 ng / μL template, 10 -2 ng / μL template, 10 - 3 ng / μL templa...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com