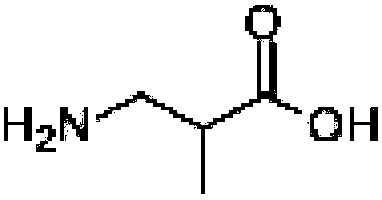
Production method for 3-aminoisobutyric acid
A technology of aminoisobutyric acid and alanine, which is applied in the field of metabolic engineering and can solve problems such as high pollution
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0046] Embodiment 1: Construction of the bacterial strain that knocks out pstG gene, fumAC gene cluster and fumB gene
[0047] 1.1 The pCAS plasmid (gifted by the Institute of Plant Physiology and Ecology, Shanghai Institutes for Biological Sciences, Chinese Academy of Sciences, see Jiang, Y., B. Chen, C. Duan, B. Sun, J. Yang and S. Yang. Applied and Environmental Microbiology. 2015,81(7):2506-2514) into Escherichia coli MG1655 by electrotransformation to obtain MG1655pCAS strain. With the pTargetF plasmid (gifted by the Institute of Plant Physiology and Ecology, Shanghai Institutes for Biological Sciences, Chinese Academy of Sciences, see Jiang, Y., B. Chen, C. Duan, B. Sun, J. Yang and S. Yang. Applied and Environmental Microbiology.2015 , 81(7):2506-2514) as a template, using pTargetFΔpstG-F / pTargetFΔpstG-R primers, carry out reverse PCR, and the above PCR products were electrotransformed into Escherichia coli DH5α competent cells (purchased from Shanghai Weidi Biotechnolo...
Embodiment 2
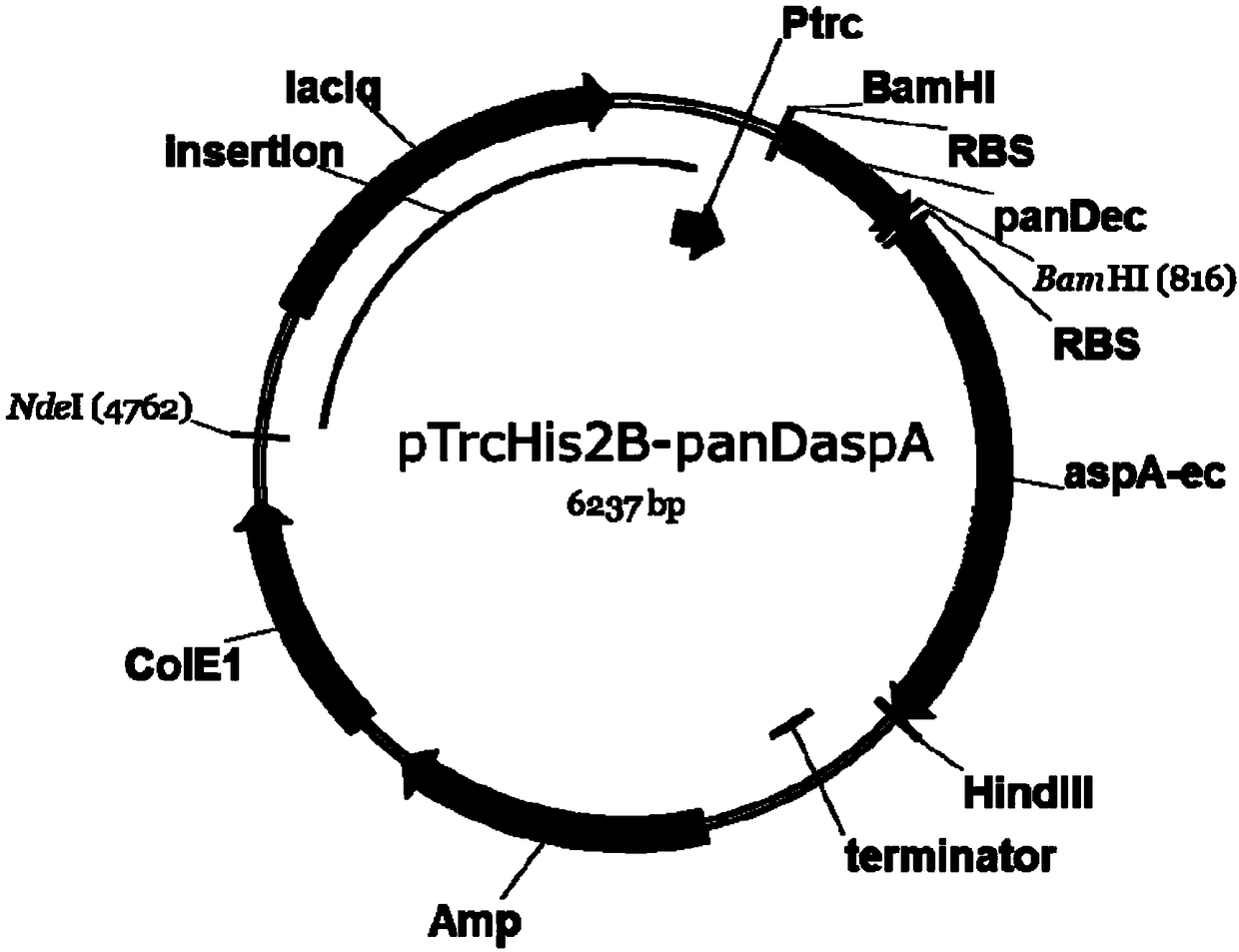
[0082] Embodiment 2: the construction of gene panD and aspA expression plasmid
[0083] The pTrcHis2B plasmid (Thermofisher Company) was digested with BamHI / HindIII to obtain the digested fragment. Using MG1655 genomic DNA as a template, PCR amplification was performed using the panD-Ec-F / PanD-Ec-R primers to obtain the panD gene fragment. Using MG1655 genomic DNA as a template, the aspA gene fragment was obtained by PCR amplification using aspA-Ec-F / aspA-Ec-R primers. PCR primer pairs are:
[0084] PanDec-F:
[0085] 5'-GGAGGAATAAACCATGGAGCTCAGGAGGTAAAAAAACATGCTGCGCACCATCCTCGG-3',
[0086] PanDec-R:
[0087] 5'-CAAATGCATTCTTAAACATGTTTTTTTACCTCTCTAAATGCTTCTCGACGTCA-3',
[0088] aspAec-F:
[0089] 5'-GACGTCGAGAAGCATTTAGAGGAGGTAAAAAAACATGTTTAAGAATGCATTTGC-3',
[0090] aspAec-R: 5'-CTGAGATGAGTTTTTGTTCTAGAATTAGTGGTTACGGATGTACT-3'.
[0091] According to Gibson The operation method of Master Mix shows that the pTrcHis2B plasmid BamHI / HindIII fragment, panD gene fragment an...
Embodiment 3
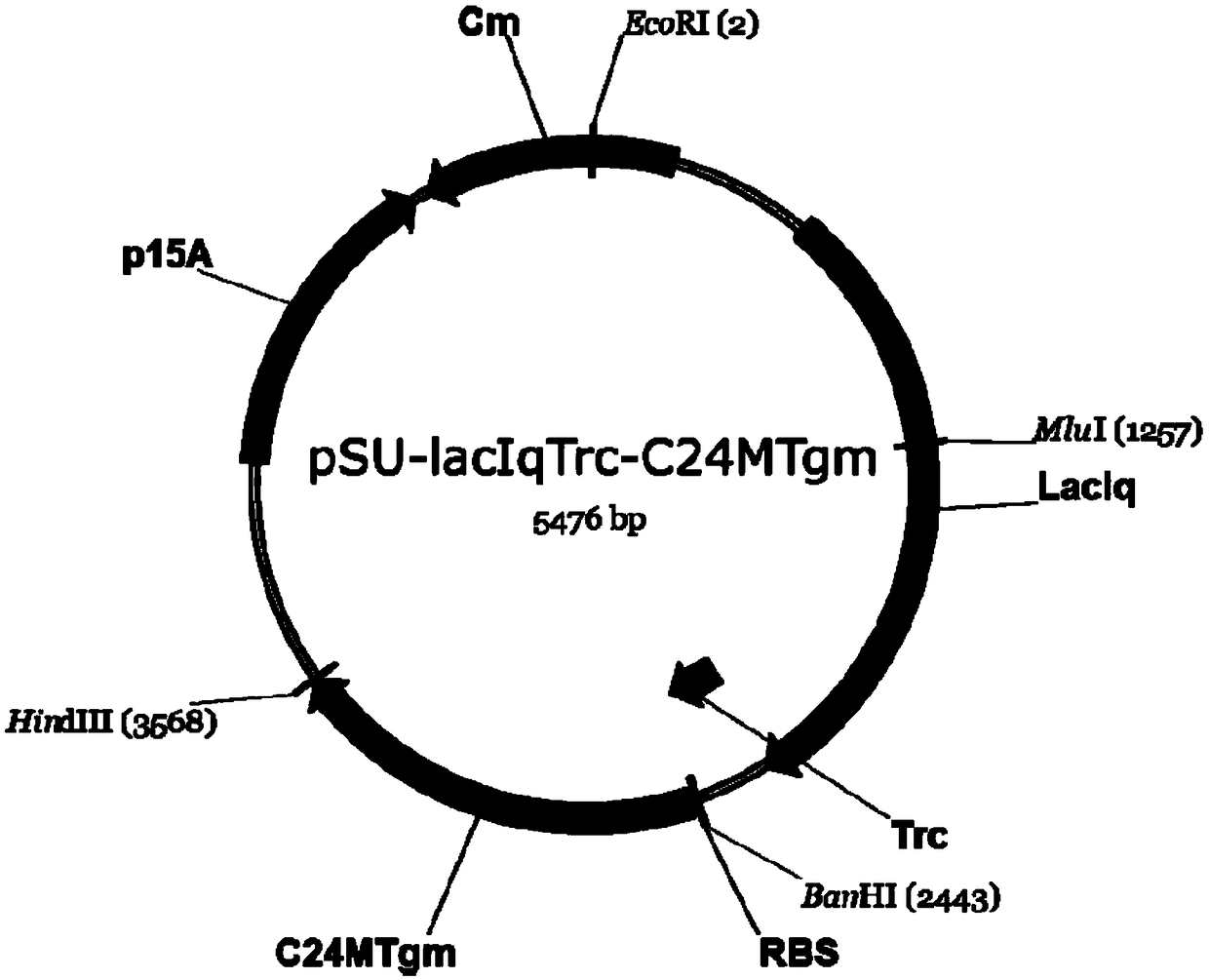
[0092] Embodiment 3: Construction of gene C24MTgm expression plasmid
[0093] 3.1 Using the pSU2718 plasmid (gifted by the Institute of Plant Physiology and Ecology, Shanghai Institutes for Biological Sciences, Chinese Academy of Sciences) as a template, use pSU2718-F / pSU2718-R primers to obtain the pSU2718 vector fragment by PCR amplification. The lacIqTrc fragment was amplified by PCR using the pTrcHis2B plasmid as a template and using lacIqTrc-F / lacIqTrc-R primers. PCR primer pairs are:
[0094] pSU2718-F: 5'-CGTAATCATGGTCATAGCTG-3',
[0095] pSU2718-R: 5'-GAATGGCGAATGGCGCTGAT-3',
[0096] lacIqTrc-F: 5'-ATCAGCGCCATTCGCCATTCGCCAGTATACACTCCGCTAT-3',
[0097] lacIqTrc-R: 5'-CAGCTATGACCATGATTACGGGAGAGCGTTCACCGACAAA-3'.
[0098] 3.2 Perform Gibson assembly and cloning of the lacIqTrc fragment and the pSU2718 vector backbone PCR fragment to obtain the plasmid pSU-lacIqTrc. S-adenosyl-L-methionine δ24-sterol-C-methyltransferase gene C24MTgm was synthesized according to NCBI'...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com