Beta-N-acetylglucosaminidase of Paenibacillus barungensis, encode gene thereof and application of beta-N-acetylglucosaminidase
A technology of glucosidase and acetylamino, which is applied in the field of Paenibacillus barengerzia β-N-acetylglucosaminidase and its coding gene and application, can solve environmental pollution and other problems, achieve good thermal stability, High specific enzyme activity and excellent enzymatic properties
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0056] Embodiment 1, the acquisition of β-N-acetylglucosaminidase and its coding gene
[0057] Extensive sequence analysis and functional verification of Paenibacillus barengoltzii, from Paenibacillus barengoltzii CAU904 (References: Zhang B., Liu Y., Yang H.Y., et al., Biochemical properties and application of a novel β-1,3-1,4-glucanase from Paenibacillus barengoltzii.Food Chemistry, 2017,234:68-75. The public can obtain from China Agricultural University) to obtain a β-N-acetylglucosaminidase The coding gene is 1038bp in full length, as shown in sequence 2 of the sequence listing. The DNA molecule shown in Sequence 2 of the Sequence Listing encodes the β-N-acetylglucosaminidase shown in Sequence 1 (composed of 345 amino acids, without signal peptide, named PbNag39).
Embodiment 2
[0058] Embodiment 2, the construction of engineering bacteria expressing β-N-acetylglucosaminidase
[0059] 1. Use the sequence 2 of the sequence listing to replace the fragment between the EcoRI and NotI restriction sites of the SUMO-pET28a vector (Novagen, Denmark) from the 1-1038 position of the 5' end to obtain the recombinant expression vector SUMO-pET28a-PbNag39( verified by sequencing).
[0060] 2. Introduce the recombinant expression vector SUMO-pET28a-PbNag39 obtained in step 1 into Escherichia coli BL21 (Bomad Gene Technology Co., Ltd.) to obtain recombinant bacteria.
[0061] 3. Introduce the SUMO-pET28a vector into Escherichia coli BL21 (Bomad Gene Technology Co., Ltd.) to obtain a recombinant bacterium converted to an empty vector.
Embodiment 3
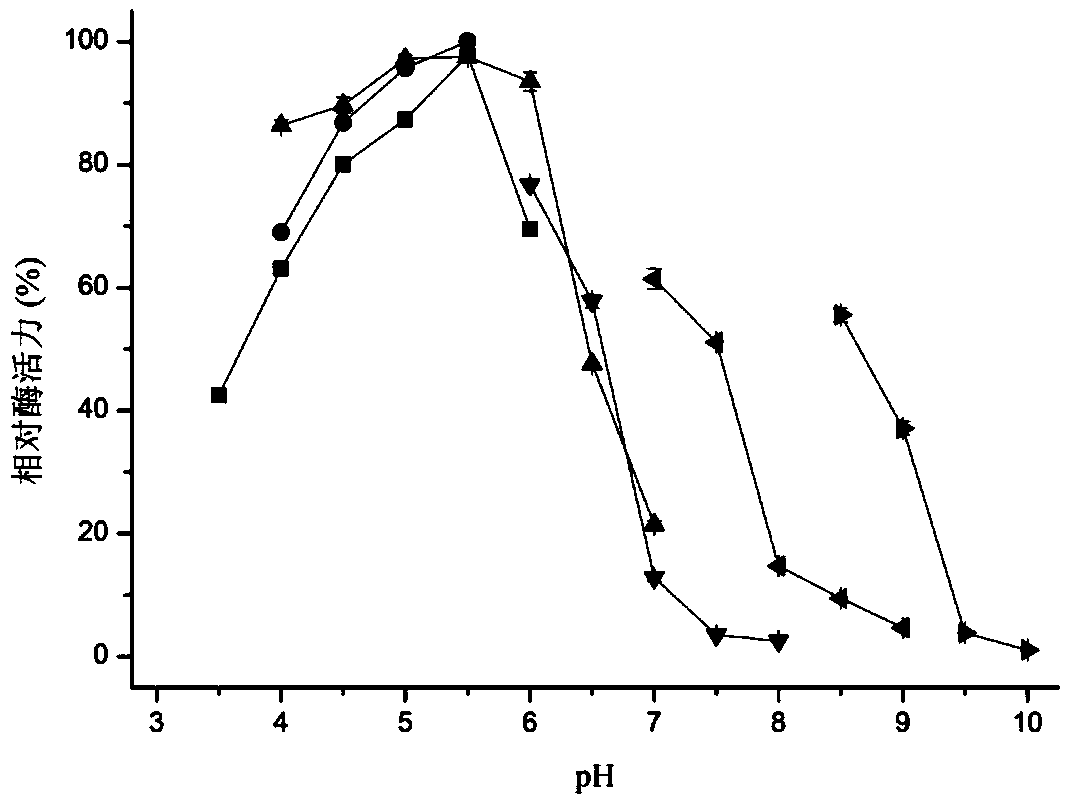
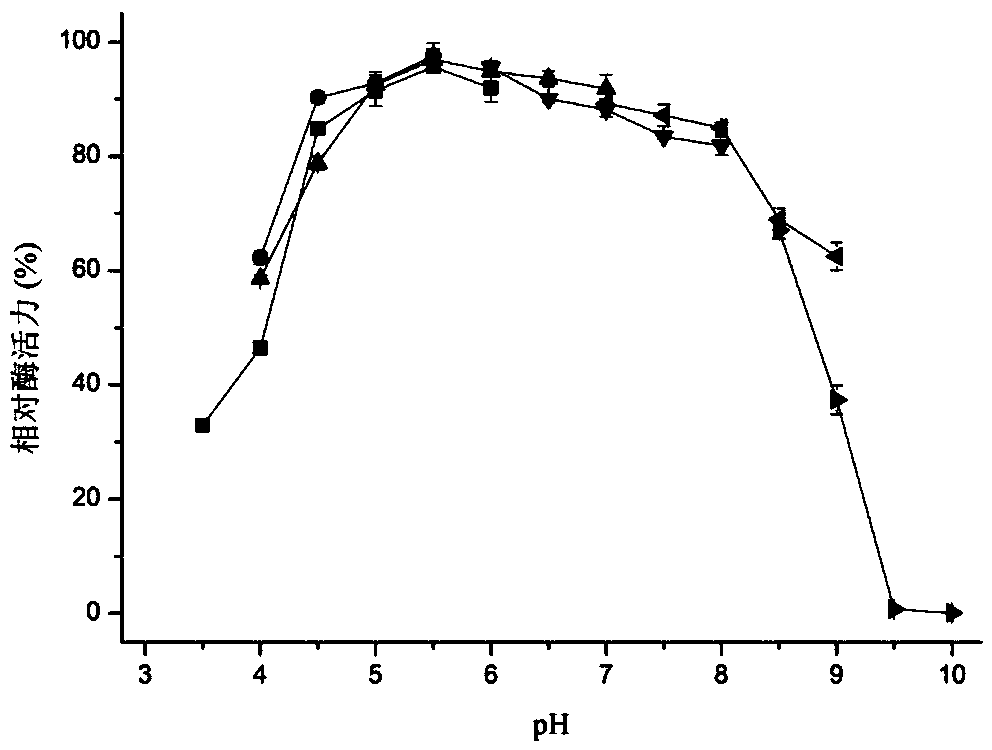
[0062] Embodiment 3, preparation of β-N-acetylglucosaminidase and detection of enzymatic properties thereof
[0063] One, the preparation of β-N-acetylglucosaminidase
[0064] 1. Inoculate the recombinant bacterium prepared in Example 2 into LB liquid medium containing 50 μg / mL kanamycin, culture it with shaking at 37°C and 200 rpm to OD 600nm Reach between 0.6-0.8, add isopropyl-β-D-thiogalactopyranoside (IPTG) in the culture system, the concentration of IPTG in the culture system is 1mmol / L, 30 ℃, 200rpm induce and cultivate overnight, then Centrifuge the culture system at 11510g, collect the bacterial precipitate, resuspend in 20mM acetic acid-sodium acetate (pH 5.5) buffer solution, and then ultrasonically break (250W, 20min). .
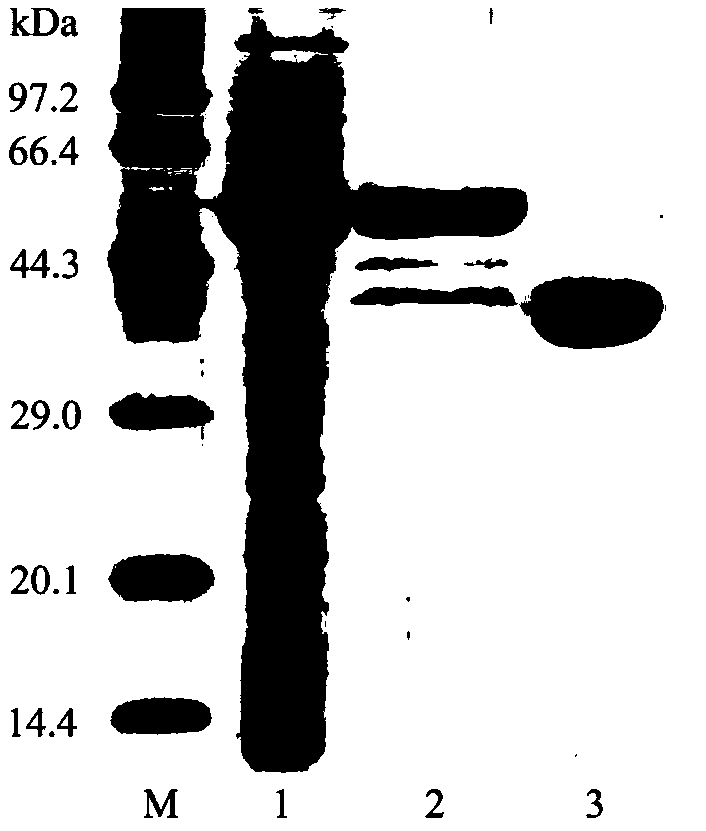
[0065] 2. Take the crude enzyme solution obtained in step 1, and use the agarose Ni Sepharose affinity column (GE Healthcare, product number: 17-5268-01) to purify the recombinant protein. The specific steps are as follows:
[0066]Equilibrate...
PUM
| Property | Measurement | Unit |
|---|---|---|
| degree of polymerization | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com