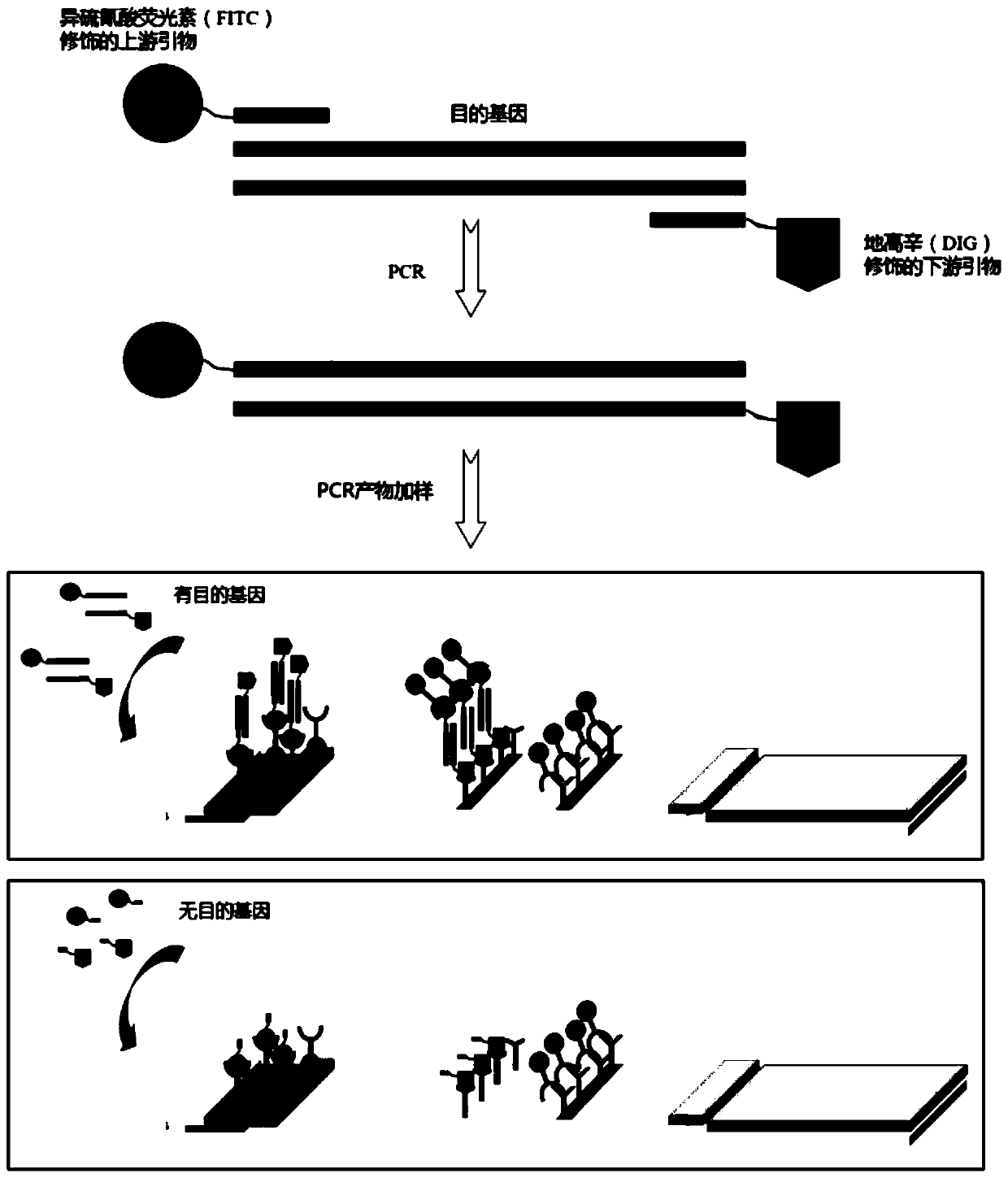
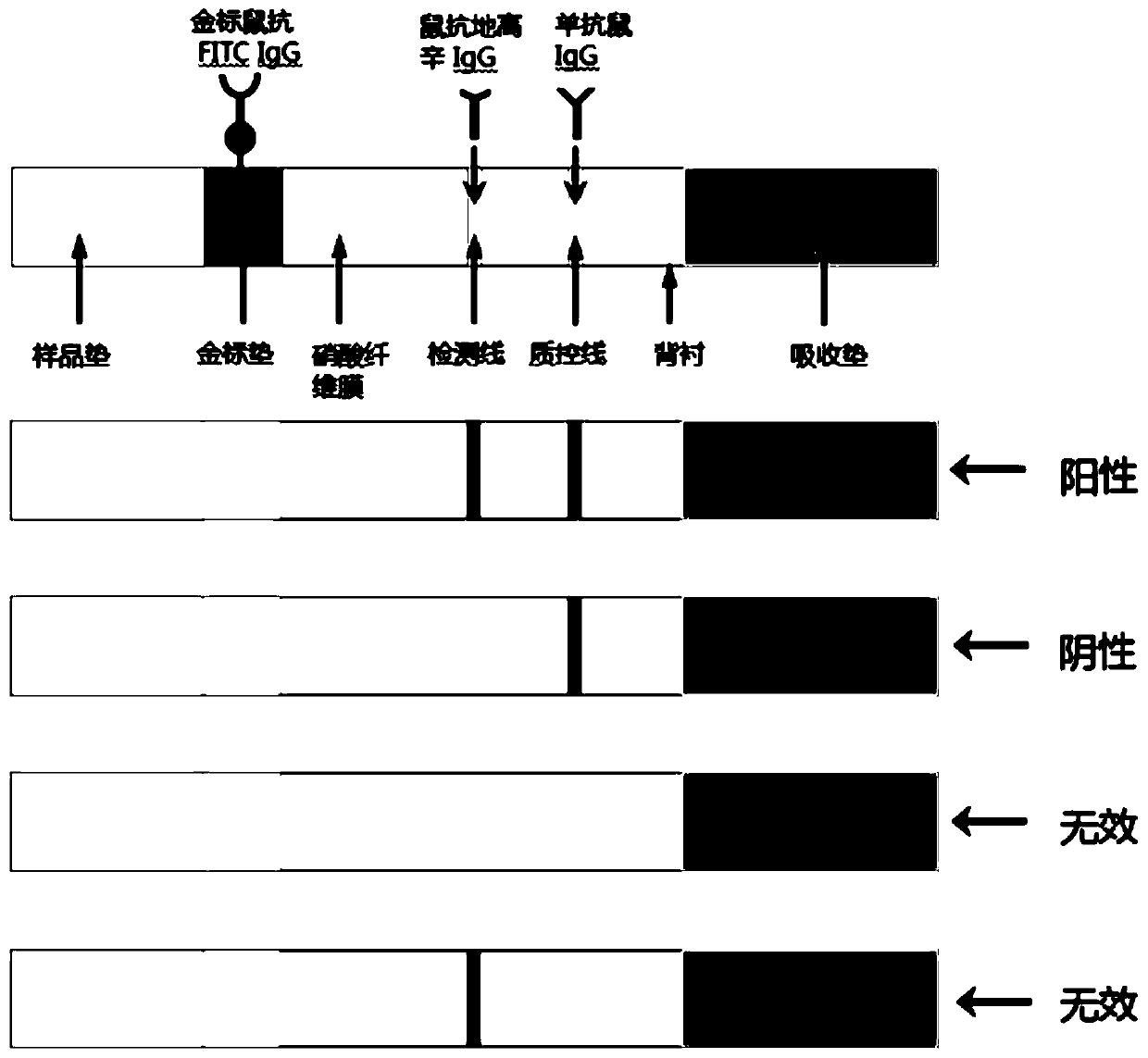
Detection reagent, kit as well as application of detection reagent and kit
A technology for detection reagents and kits, applied in the fields of molecular biology and immunology, can solve the problems of increased sensitivity and reliability, high reagent costs, complicated operations, etc., to reduce costs and convenience, improve detection sensitivity, and improve The effect of sensitivity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0096] Test the effect of different colloidal gold particle sizes on the color rendering effect of test strips.
[0097] 1 Materials and methods
[0098] 1.1 Materials
[0099] Colloidal gold solutions with different particle sizes from 10nm to 50nm are prepared according to the examples of the present invention.
[0100] 1.2 The appearance changes of gold nanoparticles with different particle sizes caused by different citric acid additions are shown in the following table:
[0101]
[0102] 1.3 PCR amplification system:
[0103]
[0104] Reaction conditions:
[0105]
[0106] 1.4 Operation method
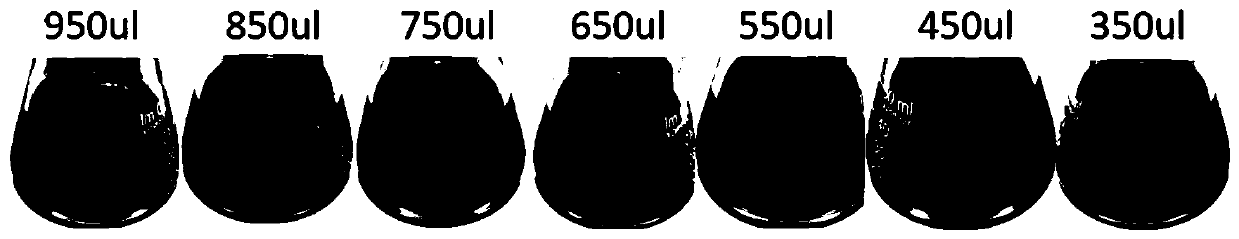
[0107] 1) Take 1ml of colloidal gold solutions with different particle sizes (as attached Figure 2a shown);
[0108] 2) Add 10ul of 1M potassium carbonate solution to 1ml of colloidal gold solution to adjust the pH of the solution, mix well; add 4ul of FITC antibody and incubate at 4 degrees for 1h; then add blocking solution and incubate at room temperature for 30m...
Embodiment 2
[0113] To test the specific identification of Escherichia coli in contamination with different foodborne pathogens.
[0114] 1 Materials and methods
[0115] 1.1 Materials
[0116] See Table 1 for various zoonotic bacterial strains involved in this example.
[0117] Table 1 Experimental strains
[0118] Table 1 Bacterial strains for detection
[0119]
[0120] 1.2 Primer design
[0121] 1.3 PCR amplification system:
[0122]
[0123]
[0124] Reaction conditions:
[0125]
[0126] 1.4 Operation method
[0127] Resuscitate cultures of different strains, count the number of colonies using plates, extract bacterial DNA, and use the above (1.3) PCR reaction system to test Escherichia coli, Staphylococcus aureus, Streptococcus agalactiae, Streptococcus dysgalactiae, Enterococcus faecalis, Grape epidermidis Coccus, yeast, Salmonella, Bacillus cereus, Listeria monocytogenes and pure water were amplified by PCR, and the specificity among different species was verifi...
Embodiment 3
[0133] To test the sensitivity of nucleic acid immunochromatography test strips to detect Escherichia coli.
[0134] 1 Materials and methods
[0135] 1.1 Materials
[0136] Escherichia coli was isolated, identified and preserved by the invention example of the School of Veterinary Medicine, Nanjing Agricultural University.
[0137] 1.2 Primer design
[0138] 1.3 PCR amplification system:
[0139]
[0140] Reaction conditions:
[0141]
[0142] 1.4 Operation method
[0143] Escherichia coli was resuscitated and cultured, and the bacteria solution was taken after gradient dilution and counted by plates. Figure 4a , 4b The number of colonies corresponding to 1-9 in the middle is 2×10 8 cfu / ml, 2×10 7 cfu / ml, 2×10 6 cfu / ml, 2×10 5 cfu / ml, 2×10 4 cfu / ml, 2×10 3 cfu / ml, 2×10 2 cfu / ml, 2×10 1 cfu / ml, 2cfu / ml; Simultaneously extract bacterial DNA as PCR template, carry out PCR amplification to Escherichia coli with above-mentioned (1.3) PCR reaction system, verif...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Sensitivity | aaaaa | aaaaa |
| Sensitivity | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com