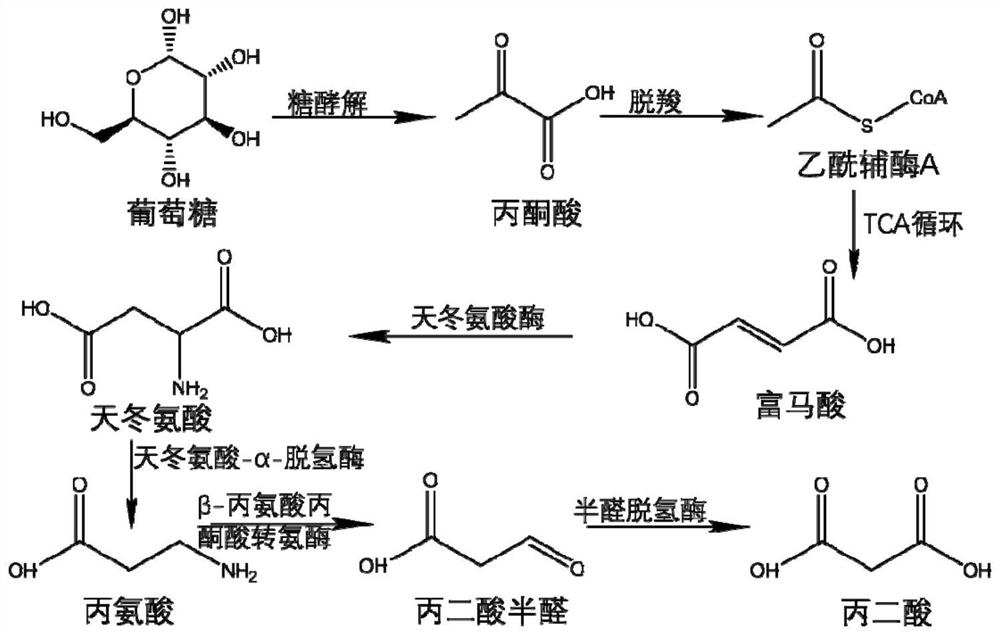
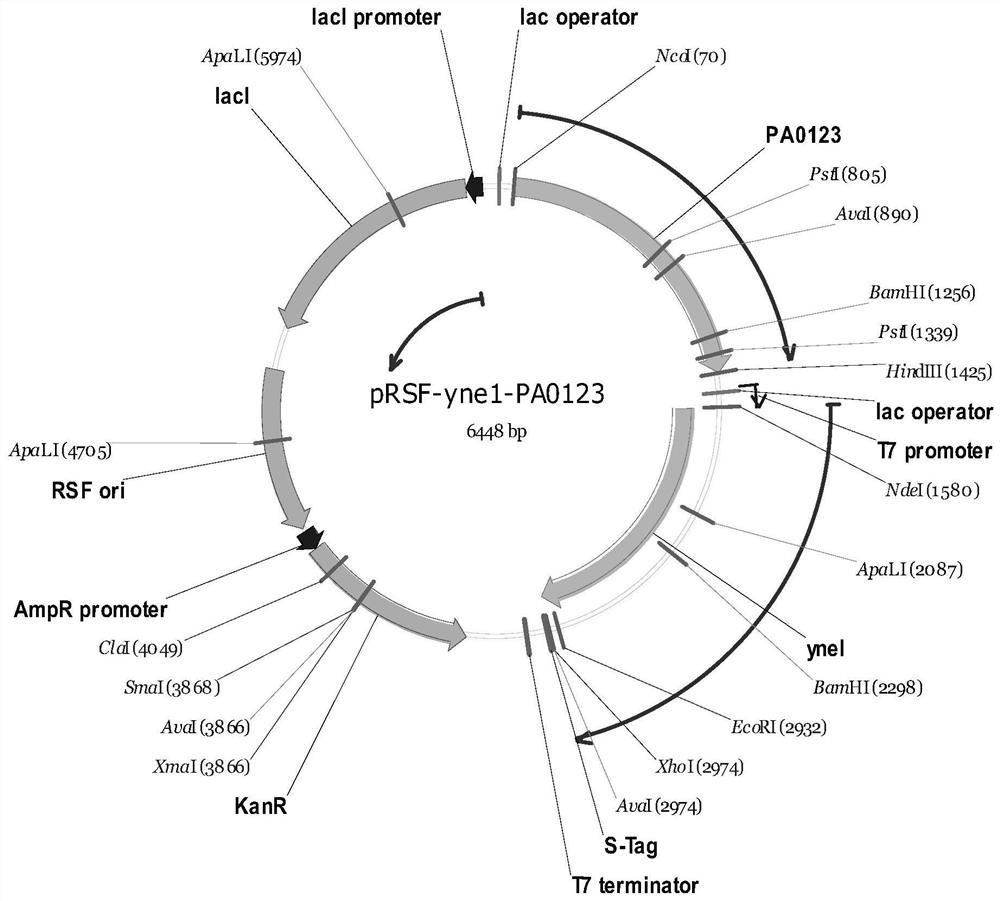
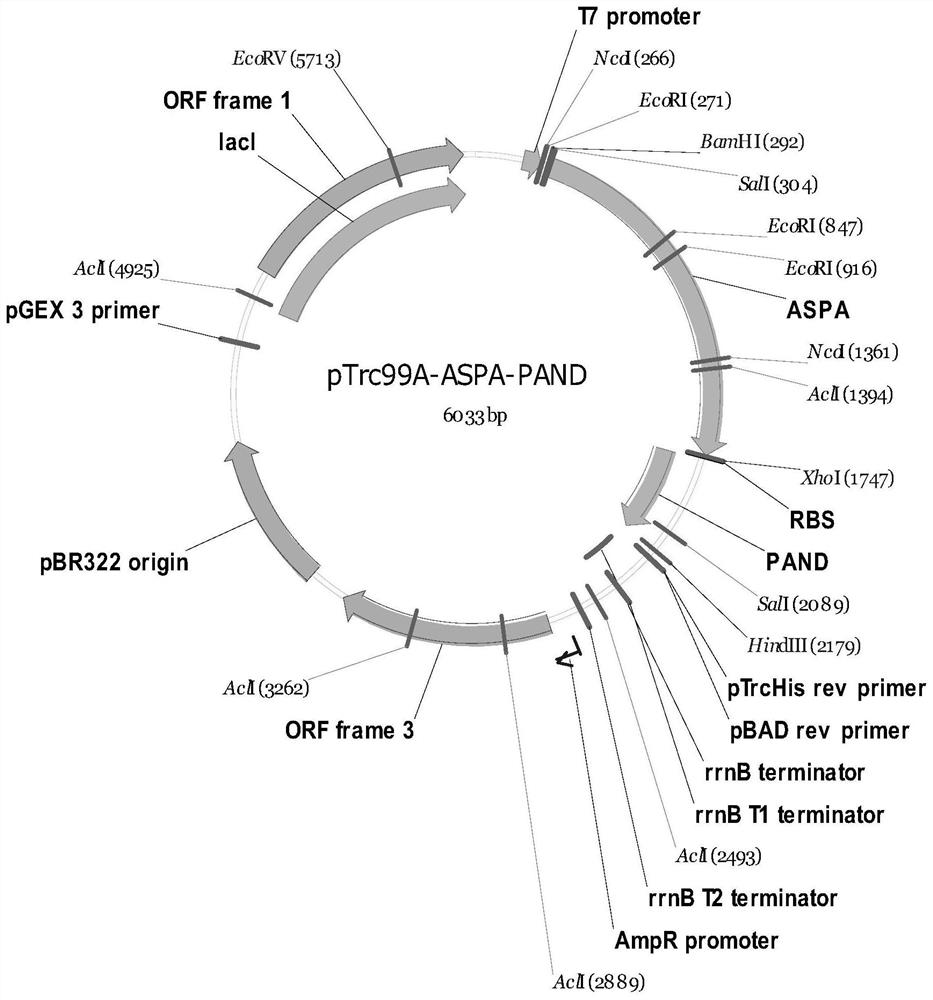
Full-biosynthesis method of malonic acid
A technology of malonate and phosphoenolpyruvate carboxylase, which is applied in the field of bioengineering, can solve the problems of difficult transformation and slow progress in the biological production of malonate, and achieve the effects of reducing pollution, low cost and simple method
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0037] Example 1: Construction of recombinant plasmids and acquisition of recombinant Escherichia coli.
[0038]The nucleotide sequences of ppc, sdhC, aspA, panD, pa0123 and yne1 are respectively shown in SEQ ID NO.1-6.
[0039] Plasmid pRSFDuet-1 was double-digested with Nco I and Hind III, and the target gene fragment pRSF-1 (3761bp) was recovered by gel cutting. The plasmid pUC57-pa0123 synthesized by Jinweizhi Company was used as a template, and primer pa0123-F / R was used to amplify by PCR. The target gene fragment pa0123 (sequence shown in SEQ ID NO.5) was obtained by increasing, and then the two target fragments pRSF-1 and pa0123 were combined with T 4 DNA ligase ligation, transfection JM109, positive transformants were picked by colony PCR, and extracted plasmids were digested and verified by PCR. The verified primers were veri-pRSF-F / R, and the verified plasmid was named pRSF-pa0123.
[0040] Plasmid pRSF-pa0123 was digested with Xho I and Nde I, and the target gene f...
Embodiment 2
[0046] Example 2: Shake flask fermentation and result analysis of recombinant Escherichia coli.
[0047] Fermentation medium: SOB medium, the composition is 20g / L tryptone, 5g / L yeast powder, 0.5g / LNaCl, 2.5mMKCl, 10mM MgCl 2 , 4g / L glucose, 50μg / ml kanamycin sulfate, 50μg / ml ampicillin, 50μg / ml streptomycin.
[0048] Seed solution preparation: Streak the strains preserved in glycerol on a plate, pick a single colony and inoculate it in a 250ml Erlenmeyer flask filled with 50ml of LB liquid medium, shake the flask overnight at 37°C and 250rpm / min.
[0049] Fermentation conditions: 2% inoculum size (1ml), inoculated in shake flask fermentation medium to make initial OD 600 0.1. Cultivate to OD at 37℃, 250r / min 600 Add 1.0mM IPTG to induce recombinant bacteria at 0.8~1.0, respectively, and change to 30°C, 250rpm / min culture.
[0050] Result analysis: During the fermentation process, samples were taken every 4 hours for the first 12 hours, and samples were taken every 12 hours...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com