Mycobacterium tuberculosis H37Rv new gene Rv2203c, and coding protein and application thereof
A technology of Mycobacterium tuberculosis, rv2203c, applied in application, genetic engineering, plant genetic improvement, etc., can solve the problems of low sensitivity, unconfirmed positive, lack of effective specific neoantigens that can identify Mycobacterium tuberculosis, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
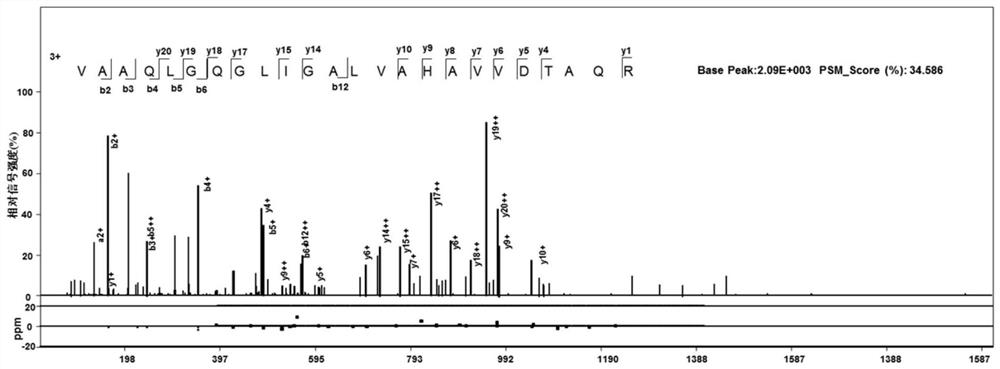
[0048] Example 1: Discovery and verification of the new gene Rv2203c (-|2468297-2469040|)
[0049] We performed a high-coverage proteome study of the Mycobacterium tuberculosis H37Rv strain using deep-coverage proteome technology. The pAnno software developed by the Institute of Computing Technology, Chinese Academy of Sciences was used to translate the whole genome (NC_000962.3) file of H37Rv published at NCBI according to the rule of translating one amino acid from three consecutive nucleotides on the positive strand and the complementary strand, using the terminator to For the translation mode of the terminator, 141,851 open reading frames (Opening reading frame, ORF) were translated using the three special initiation codons ATG, GTG and TTG of MTB. This database was used to search the H37Rv high-coverage proteome data. In order to reduce the false positive rate, we carried out strict quality control at the two levels of annotated peptides and new peptides during the data ...
Embodiment 2
[0057] Example 2: Establishing a method for identifying target gene-specific verification
[0058] (1) Design primers:
[0059] Mycobacteria sequences (nearly 200 species, 8,046 strains) that have been sequenced from the NCBI database were downloaded, and the Mycobacteria database was constructed. After comparing the local homologous sequences of the target gene, primers were designed for regions with homologous but different sequences ( image 3 ).
[0060] PCR-specific primers were designed using Primer Premier 5 to compare the difference regions of the target gene with 300 bp extensions before and after (SEQ ID NO.4). The primer sequences are as follows:
[0061] F1: 5'-TAAAACGCTAGTACGACCCCGCGGT-3' (SEQ ID NO.5);
[0062] R1: 5'-TGCCGCTCATCCCACGAACCCCAAG-3' (SEQ ID NO. 6).
[0063] The positional relationship between the above primers and the Rv2203c (-|2468297-2469040|) gene is shown below, where the subscript wavy line corresponds to the position of the primer, the bl...
Embodiment 3
[0073] Example 3: Prokaryotic cloning, expression, purification and mass spectrometry identification of Rv2203c (-|2468297-2469040|) gene
[0074] 3.1 Cloning of Rv2203c(-|2468297-2469040|) gene
[0075] (1) Design primers:
[0076] PCR primers were designed using Primer Premier 5, and EcoR I and Hind III restriction endonuclease sites were added to the upstream and downstream primers, respectively, to facilitate subsequent cloning construction. The primer sequences are as follows:
[0077] F2: 5'-GAATTCATGTGGAAGGCGCGTCGAT-3' (SEQ ID NO.7);
[0078] R2: 5'- AAGCTTTCATCCCACGAACCCCAAG-3' (SEQ ID NO. 8).
[0079] (2) The total DNA of the M.tuberculosis H37Rv strain was provided by the Tuberculosis Division of the Institute of Infectious Diseases, Chinese Center for Disease Control and Prevention.
[0080] (3) Amplify the DNA fragment, carry out polymerase chain reaction (PCR), and use the above-mentioned F2 / R2 primers to amplify.
[0081] PCR system (25μL) is ddH 2O (9.5 μL...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com