Full-biosynthesis method of pimelic acid
A technology for pimelic acid and cell production of pimelic acid, applied in the field of bioengineering, can solve the problem that the synthesis mechanism is not further elucidated, and achieve the effects of clear reaction mechanism, increased yield and optimized process
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0042] Example 1: Knockout of lactate dehydrogenase gene (ldhA), acetyl-CoA acetyltransferase gene (atoB) and succinyl-CoA synthetase α subunit gene (sucD) in BL21 (DE3)
[0043] The gene number of ldhA is ECD_01352, the gene number of atoB is ECD_02151, and the gene number of sucD is ECD_00688.
[0044] Using CRISPR / Cas9 technology, primers were designed according to the nucleotide sequence of the target gene published on the KEGG website, and the ldhA gene was knocked out. The pTarget plasmid required for the knockout was designed and provided by Suzhou Hongxun Company.
[0045] Obtaining of upstream and downstream homology arm fragments: using ldhAUP500-F, ldhAUP500-R and ldhADOWN500-F, ldhADOWN500-R as two pairs of primers, using the Escherichia coli BL21 (DE3) genome as a template, the size of A 515bp DNA fragment, which is a homologous sequence with the upstream and downstream 515bp of the ldhA gene, was recovered and purified by gel, and the purified upstream and downst...
Embodiment 2
[0052] Embodiment 2: Construction of recombinant plasmid and acquisition of recombinant Escherichia coli
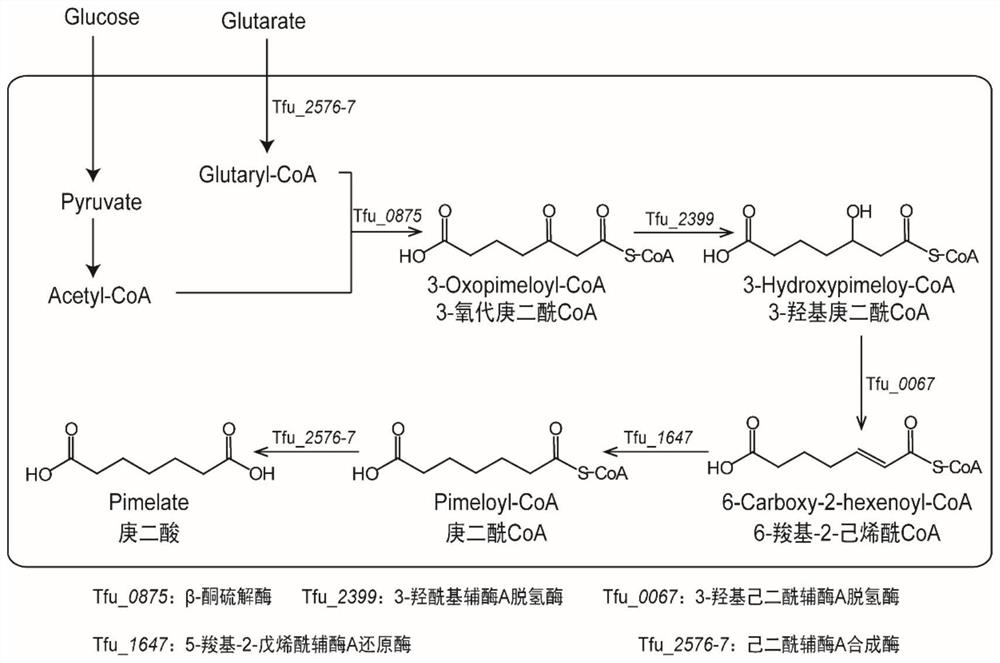
[0053] The NCBI accession number of gene Tfu_0875 is MH157180; the NCBI accession number of gene Tfu_2399 is MH157181; the NCBI accession number of gene Tfu_0067 is MH157182; the NCBI accession number of gene Tfu_1647 is MH157183; for MH157194.
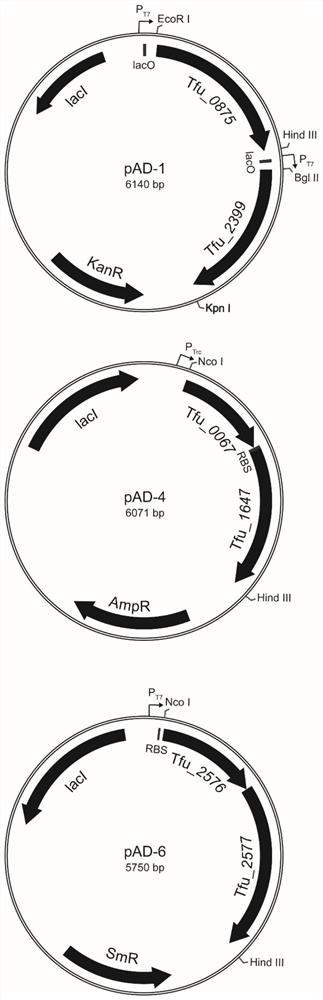
[0054] Plasmid pRSFDuet-1 was digested with EcoRI and HindⅢ, the vector fragment (3798bp) was recovered and purified by gel, and the plasmid pUC57-Tfu_0875 was digested with the same method, and the target gene fragment Tfu_0875 (shown in SEQID NO.1) was obtained by gel recovery and purification ), and then use T4 DNA ligase to connect the two fragments, transform into JM109 competent cells, pick the transformants for colony PCR verification, and extract the plasmids from the positive transformants for verification, and the verified plasmid is named pRSF-0875.
[0055] Recombinant plasmid pRSF-0875 was digested with BglⅡ and Kpn...
Embodiment 3
[0058] Embodiment 3: shake flask fermentation and result analysis of recombinant escherichia coli
[0059] Shake flask fermentation system 50mL.
[0060] Fermentation medium:
[0061] SOB medium, the composition is 5g L -1 Yeast powder, 20g· L-1 Tryptone, 5g·L -1 NaCl, 2.03g L - 1 MgCl 2 ·6H 2 O, 0.186g L -1 KCl, 4g L -1 Glucose, 6.6g·L -1 Glutaric acid, 50μg·mL -1 Kanamycin Sulfate, 50μg·mL -1 Ampicillin, 50 μg mL -1 streptomycin.
[0062] Seed liquid preparation: Streak the strains preserved in glycerol on the plate, pick a single colony and inoculate it in a triangular flask containing 50ml LB medium, 37°C, 250rpm·min -1 Shake the flask overnight. The next day, take 1ml of the bacterial solution and transfer it to 50ml of LB liquid medium, at 37°C, 250rpm·min -1 Incubate for 12-16 hours.
[0063] Fermentation conditions: the strain was inoculated in the fermentation medium at an inoculum size of 2% (2mL / 100mL), 37°C, 250rpm·min -1 Add 1mM IPTG to induce re...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com