Azoospermia chromosome variation detection kit
A mutation detection and chromosome technology, which is applied in the determination/inspection of microorganisms, DNA/RNA fragments, recombinant DNA technology, etc., can solve the problems of long cycle, many detection sites, complex process, etc., to reduce detection cost and detection sensitivity. The effect of improving and high detection sensitivity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
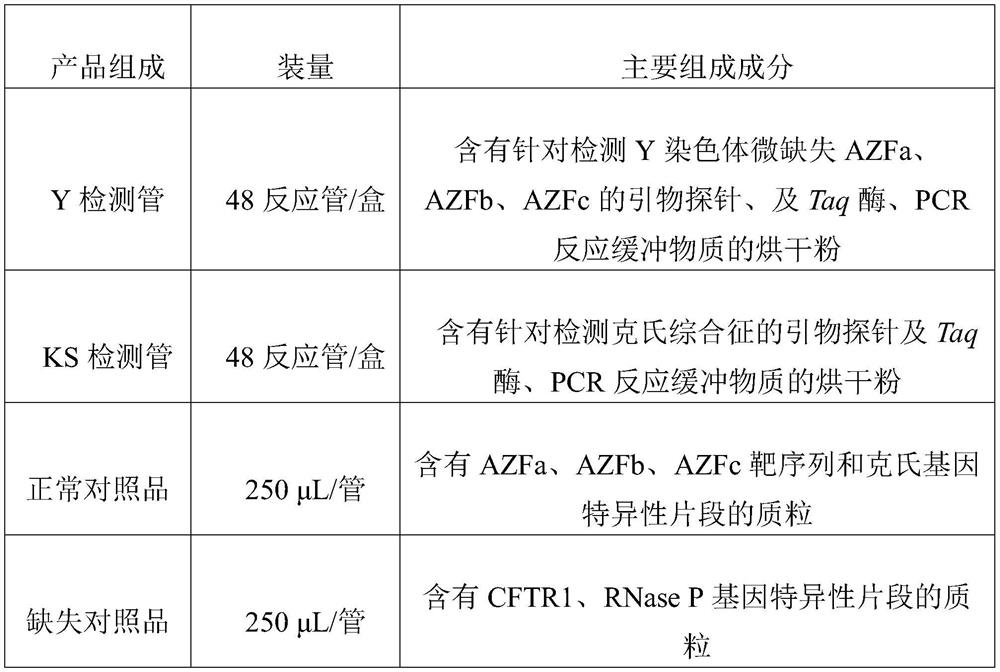
[0028] The multiple real-time fluorescent PCR method of the present embodiment, the primer probe and the kit (48 reactions / box) for detecting the chromosomal variation of azoospermia and oligospermia include components, as shown in Table 1:
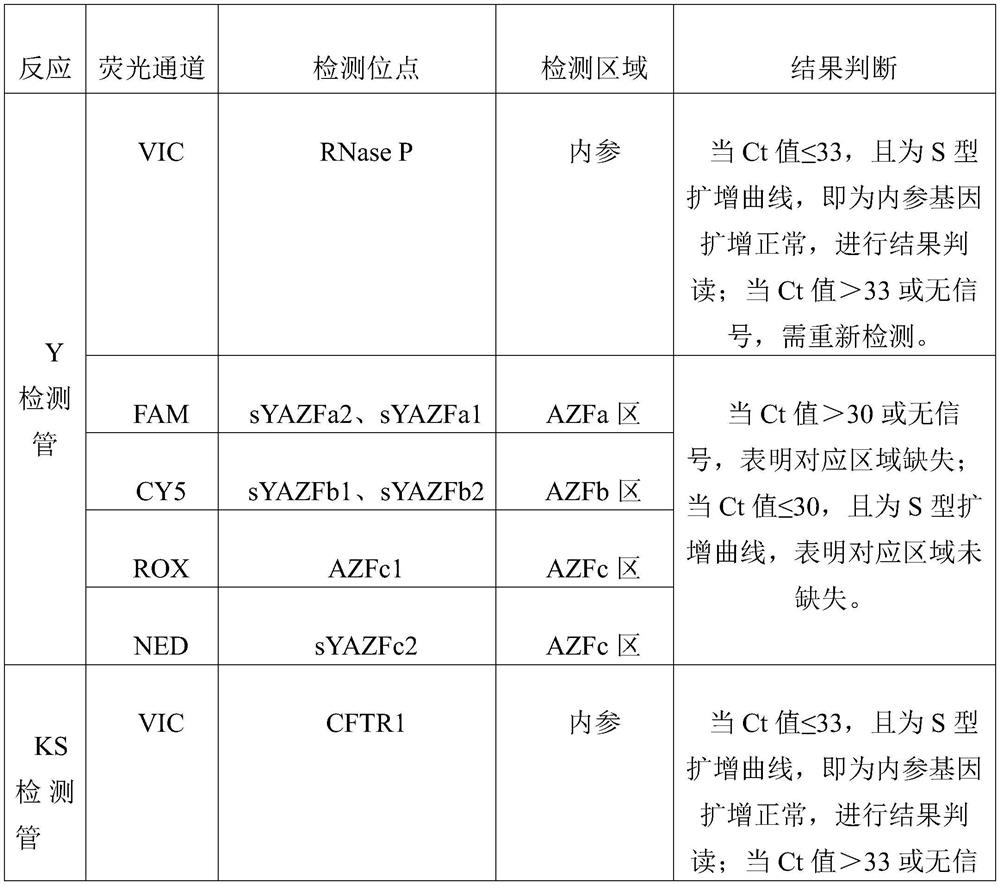
[0029] (1) Y detection tube: dry powder containing primer probes for detecting Y chromosome microdeletions AZFa, AZFb, AZFc, Taq enzyme, and PCR reaction buffer substances. The sequences of the primer probes included are:
[0030] SEQ ID NO.1: AZFa1-F: 5'-AGTGAAGGAATAACATGCCGAG-3'
[0031]SEQ ID NO.2: AZFa1-R: 5'-AAACTGTATATACCATAATCCCT-3'
[0032] SEQ ID NO.3: AZFa1-P:5'-FAM-AAGTGGTGATGGATGAGGAGT-3'-QSY
[0033] SEQ ID NO.4: AZFa2-F: 5'-CTGGGCCCAAGACACATTGT-3'
[0034] SEQ ID NO.5: AZFa2-R: 5'-ACAACTCTGGGAAGCCATTACC-3'
[0035] SEQ ID NO.6: AZFa2-P:5'-FAM-CCATGGATCTCACTTTGCAGGACAGAGAC-3'-QSY
[0036] SEQ ID NO.7: AZFb1-F:5'-TATAGCCCAAAACTAATCAGCATC-3'
[0037] SEQ ID NO.8: AZFb1-R: 5'-TGGTAGATTCCAGTGGGTGCTATC-3'
[0038] SEQ ID NO....
Embodiment 2
[0066] The method for real-time fluorescent quantitative PCR detection of human azoospermia and oligospermia chromosomal variation using the kit described in Example 1:
[0067] (1) Nucleic acid extraction:
[0068] Take 200 μL whole blood in vitro sample, use Tianlong automatic nucleic acid extractor (NP968-3S) and matching Tianlong whole blood genomic DNA extraction kit to extract the whole blood in vitro sample collected by EDTA anticoagulant tube, and use Micro-volume ultraviolet spectrophotometer to determine the purity and concentration of nucleic acid, its OD 260 / 280 Between 1.6-2.0; dilute the genomic DNA concentration to 5ng / μL with sterile double distilled water for later use.
[0069] (2) In vitro sample detection:
[0070] Add the absence control substance, normal control substance, and the isolated sample DNA to be tested into the reaction wells of the Y detection tube and the KS detection tube, respectively, with a sample volume of 20 μL.
[0071] (3) Amplific...
Embodiment 3
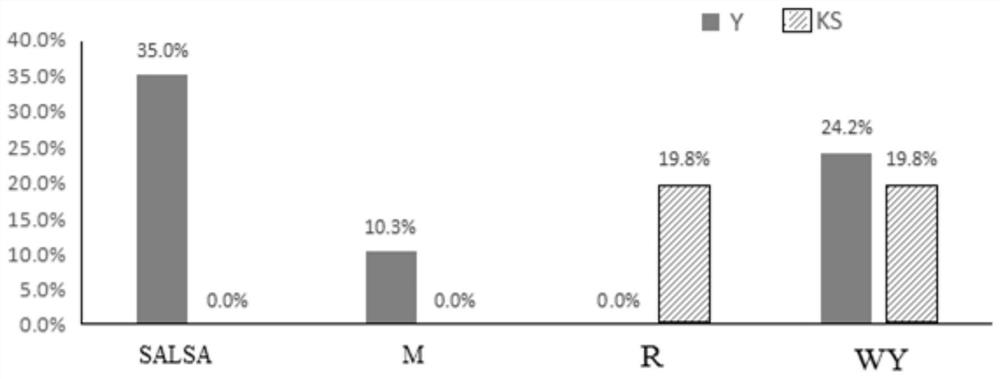
[0096]Two negative samples were tested using the kit described in Example 1 and a commercially available Y chromosome microdeletion detection kit (fluorescent PCR method). The DNA concentration used was serially diluted to 20ng / μL, 10ng / μL, 5ng / μL, 2ng / μL, 1ng / μL respectively, and the Ct value of the internal reference signal was detected. The internal reference Ct values of the two kits are as follows:
[0097] Table 6 Comparison of the detection limit of the internal reference signal between this kit and a commercially available Y staining kit
[0098]
[0099] It can be seen from the above results that after the drying process is adopted in this kit, the total sample consumption is greatly reduced, and the sensitivity is 5 times higher than that of the traditional fluorescence quantitative kit.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com