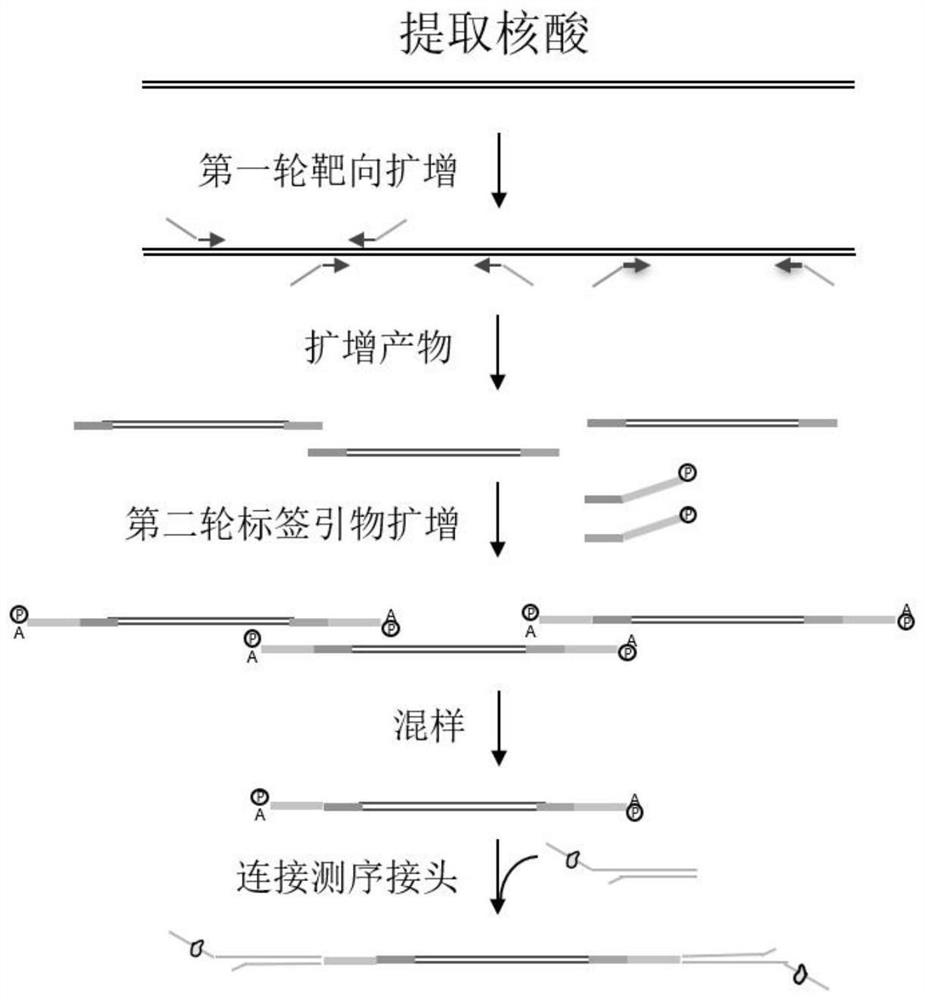
Rapid pathogenic microorganism detection method and kit thereof
A technology of pathogenic microorganisms and detection methods, applied in the field of rapid detection methods and kits for pathogenic microorganisms, can solve the problems of difficult interpretation of sequencing results, long sequencing time, and short sequencing read length, etc., and achieve the effect of improving the success rate of library construction
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
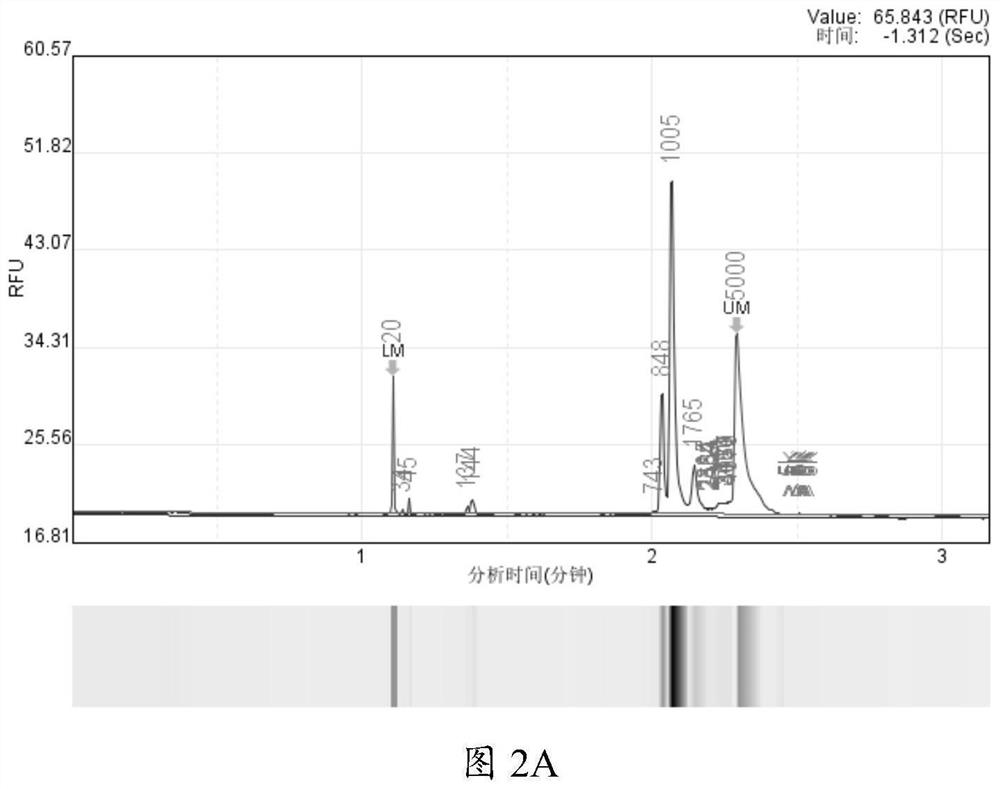
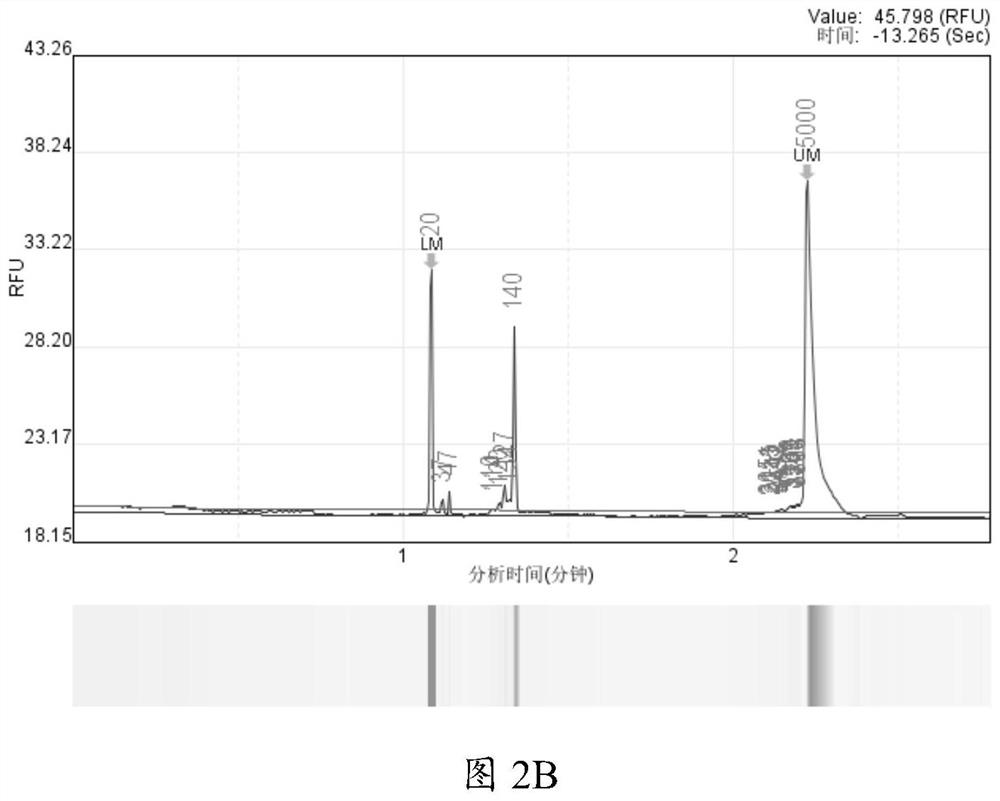
[0065] Embodiment 1 Comparison test between the present invention and Nanopore official process
[0066] For the database construction technology, Nanopore official also provided part of the experimental process. In order to compare the difference between the experimental results of the detection method of the present invention and the official Nanopore process, a comparative test was carried out using standard products. The main comparative test conditions are as follows:
[0067] Table 6 Comparison of test conditions between the method of the present invention and the Nanopore official process
[0068] Experimental items official process this invention Nucleic acid amount 30ng 50pg~100ng Amplification process One Tube Direct Expansion two-stage amplification connection method ligase-independent ligation ligase ligation
[0069] When using the DNA extracted from a single cultured microorganism, according to the required 30ng test, ...
Embodiment 2
[0074] Embodiment 2 The inventive method standard substance detection limit test
[0075] For clinical applications, the detection limit is a key indicator for judging the experimental effect. The currently known methods require the number of microorganisms to be extracted to reach 20 or more copies in order to be detected normally. For standard microbial strains, the following 20 , 10, and 4 copy number detection tests, the specific detection results are as follows:
[0076] Table 8 standard detection limit experimental results
[0077]
[0078]
[0079] Microbiological standards are microbial solutions diluted to 100 copies / mL, including Candida albicans, Staphylococcus aureus, Staphylococcus epidermidis, Escherichia coli, Enterococcus faecalis, Acinetobacter baumannii, Candida glabrata, EBV virus and CMV For viruses, 200uL, 100uL, and 40uL of standard bacterial solutions were extracted respectively during nucleic acid extraction. After extraction, pathogenic microorg...
Embodiment 3
[0080] Embodiment 3 clinical blood sample detection
[0081] Take 10 cases of whole blood samples with clear pathogenic microorganisms in clinical blood culture, take 200uL whole blood to extract nucleic acid, and then perform library construction and sequencing according to the method of the present invention. All 10 cases of samples were normally built and sequence data were obtained. The specific detection results are as follows :
[0082] Table 9 The detection results of different clinical blood samples
[0083] sample blood culture results Is it consistent blood 1 Escherichia coli unanimous blood 2 Staphylococcus epidermidis not detected blood 3 Pneumonia Kleb unanimous blood 4 Staphylococcus haemolyticus unanimous blood 5 Staphylococcus aureus unanimous blood 6 Enterococcus faecium unanimous blood 7 Pseudomonas aeruginosa unanimous blood 8 Streptococcus pneumoniae unanimous blood 9...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com