Genetically engineered bacterium for producing fucosyllactose and production method
A technology of fucosyllactose and genetically engineered bacteria, applied in the fields of metabolic engineering and food fermentation, can solve the problem of low yield of fucosyllactose, and achieve the effects of cheap substrate, fast growth of strains, and efficient synthesis
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
preparation example Construction
[0056] Preparation of Escherichia coli Competent: Kit of Shanghai Sangon Bioengineering Company.
[0057] LB liquid medium: 10g / L peptone, 5g / L yeast extract, 10g / L sodium chloride.
[0058] LB solid medium: 10g / L peptone, 5g / L yeast extract, 10g / L sodium chloride, 18g / L agar powder.
[0059] Fermentation medium: glycerol 30g / L, potassium dihydrogen phosphate 13.5g / L, citric acid 1.7g / L, diammonium hydrogen phosphate 4.0g / L, magnesium sulfate heptahydrate 1.4g / L, yeast extract 10g / L, Trace metal solution 10mL / L (ferric citrate 10g / L, magnesium sulfate heptahydrate 2.25g / L, copper sulfate pentahydrate 1.0g / L, manganese sulfate monohydrate 0.35g / L, borax 0.23g / L, molybdic acid Ammonium 0.11g / L, calcium chloride dihydrate 2.0g / L), pH 6.8.
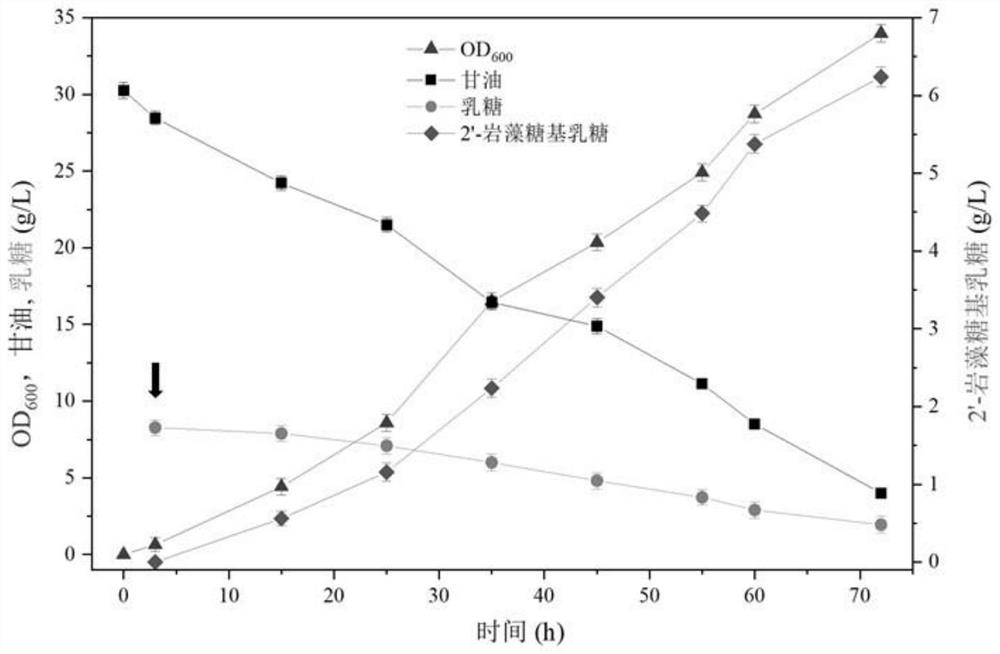
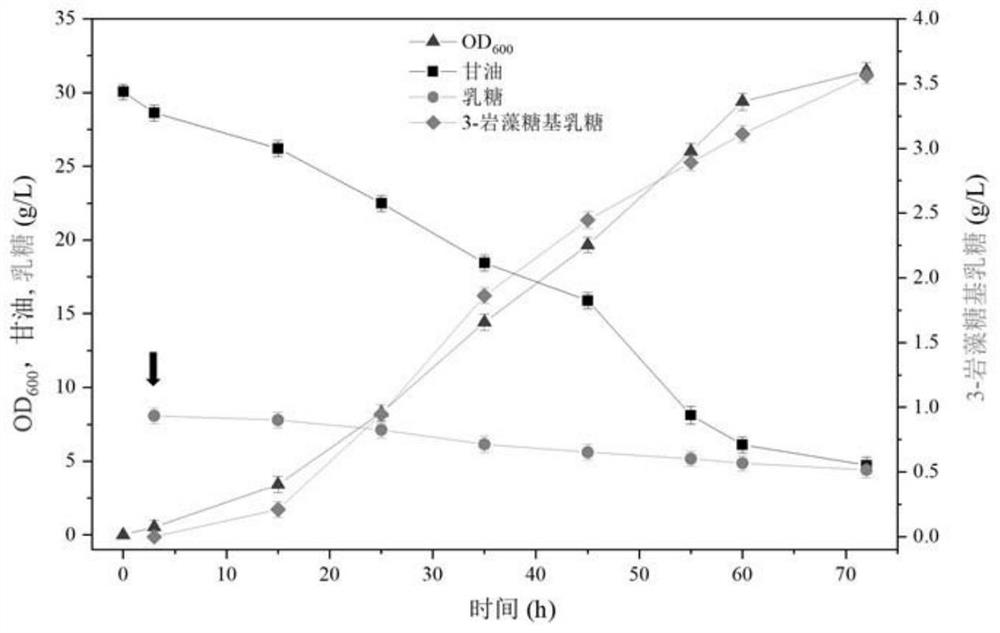
[0060] Determination of 2'-FL and 3-FL:
[0061] HPLC measurement: 1 mL of fermentation broth was boiled at 100°C for 10 min, centrifuged at 12000 r / min for 5 min, the supernatant was filtered through a 0.22 μm membrane, and the production ...
Embodiment 1
[0063] Example 1: Transformation of Escherichia coli chassis microorganisms by CRISPR-Cas9 gene knockout technology
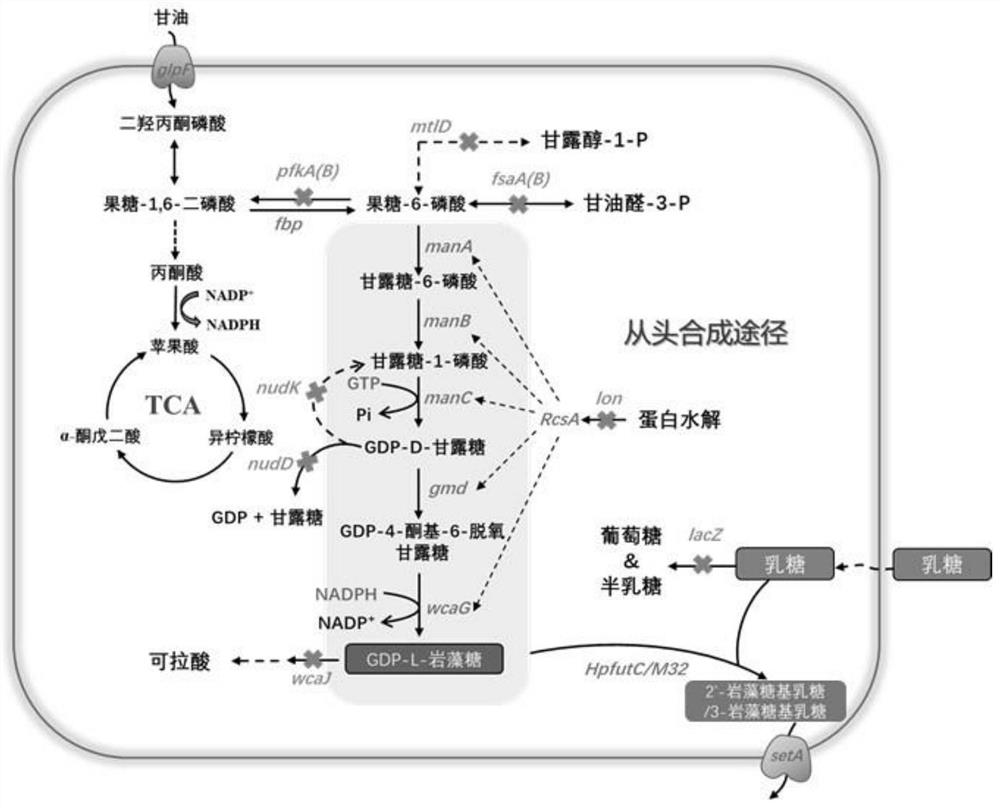
[0064] According to the metabolic pathway of fucosyllactose ( figure 1 shown), using Escherichia coli BL21(DE3)ΔlacZΔwcaJ as the starting strain (the construction method of Escherichia coli BL21(DE3)ΔlacZΔwcaJ has been disclosed in the patent literature with publication number CN112342176A), using the CRISPR-Cas9 gene knockout system separately or simultaneously The nudD, nudK, pfkA, pfkB, mtlD, lon, fsaA, fsaB, and fucIK genes in the genome of Escherichia coli BL21(DE3)ΔlacZΔwcaJ were knocked out, and the specific steps were as follows (see Table 1 for the primer sequences involved):
[0065] (1) Take the nudD gene as an example, find the specific target gRNA (20bp) of the target gene through http: / / www.regenome.net / cas-offinder, use nudD-gRNA-F / gRNA-R upstream and downstream primers, PCR amplification was performed using pTargetF plasmid (Addgene: #62226) as...
Embodiment 2
[0076] Example 2: Construction of de novo synthesis pathway and batch fermentation of knockout strains to produce fucosyllactose
[0077] The specific steps for the construction of the expression vector for the de novo synthesis pathway are as follows (see Table 2 for the primer sequences involved):
[0078] (1) Obtaining manB, manC, gmd and wcaG fragments: using the genome of Escherichia coli K12 as a template, using primers manCB_F / R and GW_F / R for PCR amplification, gel recovery DNA, and obtaining manC-manB, gmd-wcaG genes fragment.
[0079] (2) Using pRSFDuet-1, pETDuet-1, pCDFDuet-1, pACYCDuet-1 and pCOLADuet-1 as templates, use primers V1_F / R and V2_F / R to amplify the vector backbone sequence. According to the In-Fusion cloning technique, the fragments manC-manB were respectively inserted between the Nco I / BamH I restriction sites of the above-mentioned vector, and the fragment gmd-wcaG were respectively inserted between the NdeI / Xho I restriction sites of the above-men...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com