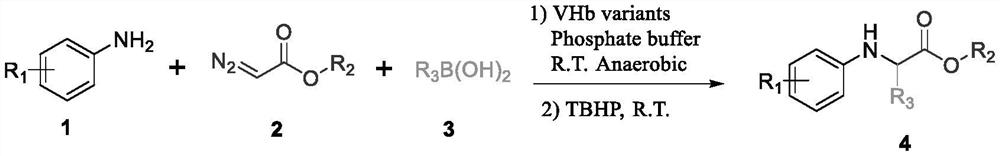
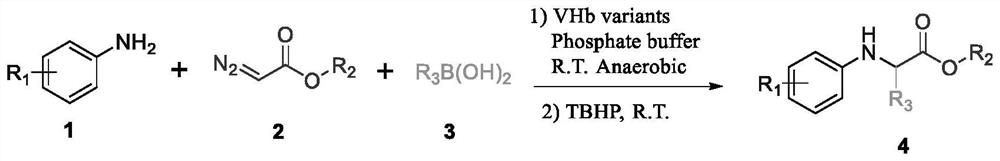
Method for catalyzing non-natural amino acid derivative by bifunctional heme protein
A technology of unnatural amino acid and heme protein, which is applied in the field of biocatalytic synthesis, can solve the problem that the number of species or catalytic performance cannot meet the needs of efficient and economical preparation of unnatural amino acids
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0025] Example 1 Construction of wild-type VHb gene
[0026] The upstream and downstream primers of VHb were designed, and the target gene was obtained by PCR technology with wild-type VHb (SEQ ID NO. 1) as the template.
[0027] Upstream primer VHb-F:
[0028]
[0029] Downstream primer VHb-R:
[0030]
[0031] The PCR reaction system is as follows:
[0032]
[0033] Reaction conditions: 95℃2min→(95℃15s→65℃15s→72℃90s)×30→72℃10min,;
[0034] After the PCR amplification product was detected by agarose gel electrophoresis, under the UV detector, cut out the target band and recover the target fragment according to the operation instructions of the gel recovery kit.
[0035] Establish a double enzyme digestion system
[0036] The vector PUC19 plasmid and the VHb target fragment were double digested with restriction endonucleases Hind III and EcoR I, respectively. The restriction enzyme digestion systems of plasmid and VHb target fragments are as follows:
[0037] ...
Embodiment 3
[0058] Example 3 Construction of random mutation library
[0059] The gene of VHbDF1 was mutated using error-prone PCR mutagenesis technique.
[0060] Using the recombinant plasmid PUC19-VHbDF1 linked with the VHbDF1 gene as a template, a pair of upstream and downstream primers containing the 19bp plasmid homology arm of the VHbDF1 gene were designed, and ep-PCR was performed to obtain the target gene fragment mutation library.
[0061] Upstream primer TYVGB-F: GGGCCATAATAATGAACTTAAGGAAGACCCTC
[0062] Downstream primer TYVGB-R: CGTTGTAAAACGACGGCCAGTGAATTCTTA
[0063] The PCR reaction system is as follows:
[0064]
[0065]
[0066] PCR program:
[0067] 95℃2min→(95℃15s→65℃15s→72℃2min)×35→72℃10min
[0068] After the PCR amplification product was detected by agarose gel electrophoresis, under the UV detector, cut out the target band and recover the target fragment according to the operation instructions of the gel recovery kit.
[0069] Establish a double enzyme dige...
Embodiment 4V
[0079] Example 4 Expression and Screening of VHb Mutation Library
[0080] The cells were cultured and passaged at 37°C at 200rpm in a 2mL×96 deep-well plate, and expressed anaerobically at 25°C at 120rpm for 30h after 24h. The plate was centrifuged at 4000 rpm for 10 min, the supernatant was discarded, and the pellet was resuspended with 180 μl PBS buffer.
[0081] Orifice plate primary sieve
[0082] Dissolve ethyl diazoacetate and aniline in DMSO at a concentration of 200 mM, add 20 μl to each well, and react at room temperature at 300 rpm for 2 hours, then add 300 mM isopropylboronic acid solution in DMSO to each well, and continue the second step for 6 hours. After the reaction, 200 μl of n-hexane: ethyl acetate = 3: 2 solvent was added to each hole for extraction, and the upper filter membrane was taken and placed in a liquid-phase sample bottle for liquid-phase high-throughput detection (n-hexane: isopropanol = 95 : 5), and screened 20 mutants with higher yields for r...
PUM
 Login to view more
Login to view more Abstract
Description
Claims
Application Information
 Login to view more
Login to view more - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap