Function identification of promoter of source sequence of gluconacetobacter xylinum and application of promoter in promotion of synthesis of bacterial cellulose
A technology of gluconacetobacter and bacterial cellulose, applied in the biological field, can solve problems such as unclear functions, high cost of inducers, and inability to open constitutive promoters, so as to promote the synthesis of d bacterial cellulose, increase cognition, Effects of high promoter activity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
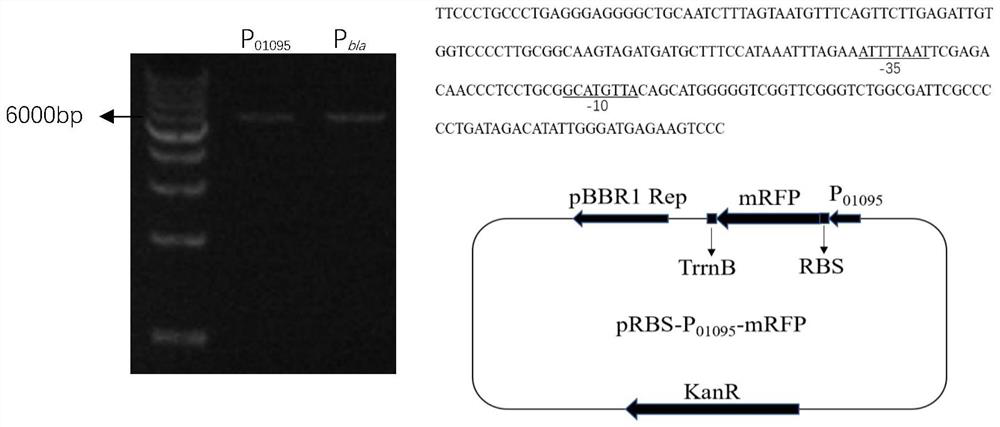
[0048] The promoter strength of the Acetobacter xylinum derived sequence characterizes the construction of the plasmid.
[0049] In order to characterize the expression strength of the promoter of the sequence derived from Acetobacter xylinum 01095 Express the red fluorescent protein gene. First, a backbone plasmid pBRS was constructed for subsequent studies. The backbone plasmid is based on the pBla vector with the promoter, ribosome binding site (RBS) and terminator removed, and is composed of BamHI restriction site, RBS, mRFP and TrrnB terminator. According to the published genome sequence of Acetobacter xylinum CGMCC 2955 and P 01095 Promoter annotation information, design primers P 01095 -F / R, using the CGMCC 2955 genome as a template to obtain P by PCR amplification 01095 promoter fragment. Use BamHI to digest the vector pRBS to obtain a linear vector, and then clone and connect the above two fragments through Novozan's one-step recombination kit to obtain the pRBS-...
Embodiment 2
[0053] Construction and strength characterization of promoter-screening plasmids for sequences derived from Acetobacter xylinum.
[0054] The red fluorescent protein gene mRFP was used as the reporter gene, and the control was P on the commonly used pBla plasmid. bla Promoter, examine expression element P 01095 Fluorescent protein expression characteristics, and its fluorescence intensity at 96h was determined.
[0055] 1. Determination of fluorescent protein expression intensity
[0056] The fluorescent protein expression assay method is as follows: pick a single colony from the solid medium plate with an inoculation loop and inoculate it in 5 mL of liquid medium at 180 r / min at 30 °C for 24 h. Transfer 1 mL of fermentation broth to 75 mL of seed medium containing 4‰(v / v) cellulase (Celluclast 1.5L, Novozymes), and cultivate to OD at 180 r / min at 30 °C. 600 is 0.5 to 0.6. Take 750μl of seed solution and transfer it to 75mL liquid medium (initial pH 6.0, initial glucose co...
Embodiment 3
[0063] Sequences with strong promoter functions derived from Acetobacter xylinum are used in the synthesis of bacterial cellulose in Acetobacter xylinum.
[0064] 1. Construction of a recombinant vector of a sequence with strong promoter function derived from Gluconobacter xylinum in Acetobacter xylinum.
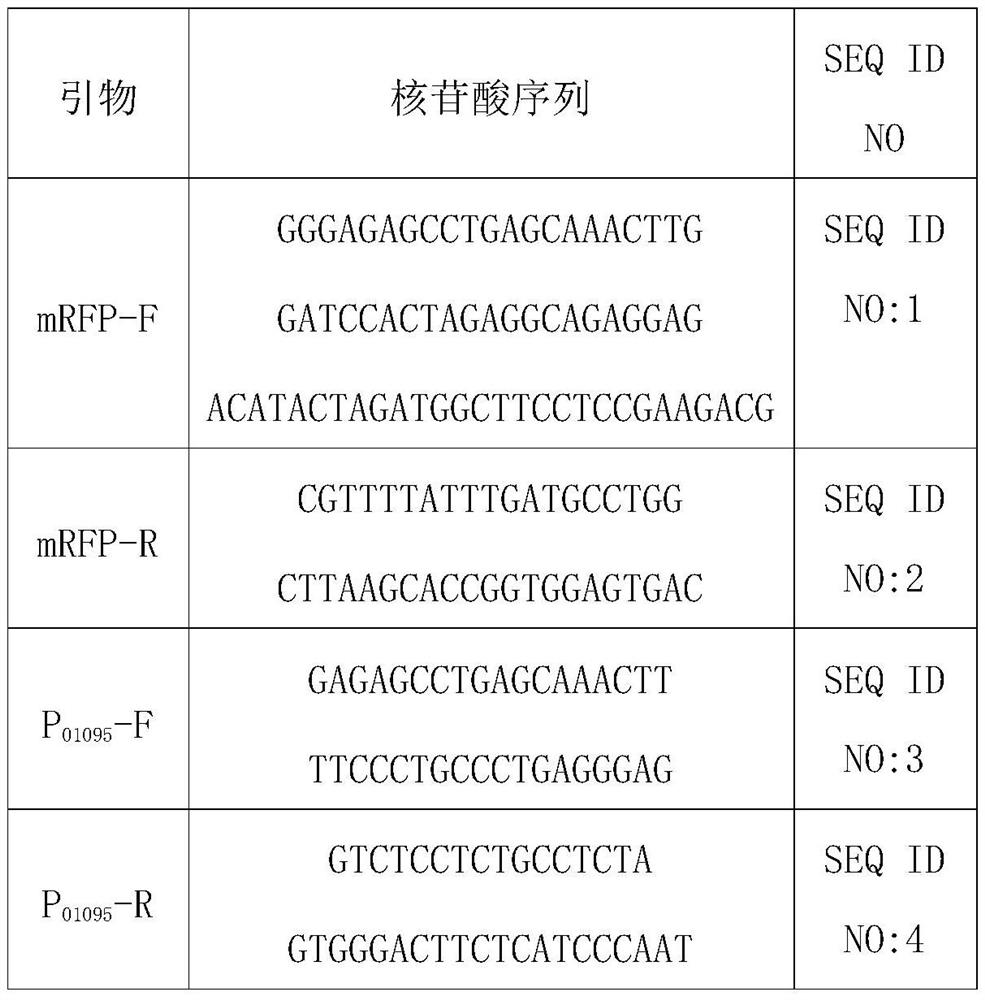
[0065] According to the reported genome sequence of Acetobacter xylinum CGMCC 2955, the genome of CGMCC2955 was used as the template, and P 01095 -pfkA-F / R,P bla - pfkA-F / R is the primer, PCR amplification of the target gene pfkA, after the fragment is recovered, cloned and ligated by Novozan's one-step recombination kit to obtain the recombinant vector pRBS-P 01095 -pfkA and pRBS-P bla -pfkA. The primer sequences used above are shown in Table 3.
[0066] table 3
[0067]
[0068] 2. Construction of bacterial cellulose-producing bacteria having a sequence with strong promoter function derived from Acetobacter xylinum.
[0069] The recombinant vector pRBS-P construct...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com