Ribotide sequence of 2-naphthanic acid degradation bacteria DNA segment and its preparation method and application
A technology of nucleotide sequence and naphthalene acid-degrading bacteria, which is applied in the field of nucleotide sequence of DNA fragments of 2-naphthoic acid-degrading bacteria, and can solve the problems that the effect has never been reported.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0019] Example 1: Extraction process of 2-naphthoic acid degrading bacteria total DNA
[0020] Inoculate 2-naphthoic acid-degrading bacteria (Burkholderia sp. JT1500) into 5ml LB medium, culture overnight at 30°C on a 140rpm shaker, centrifuge at 6000g for 10min, and collect the bacteria. Add 2.7ml DNA extraction buffer, fully suspend; add 20μl 20mg / ml proteinase K, shake at 200rpm in a 37°C water bath shaker for 30min, then add 0.3ml 20% SDS, gently invert several times to mix well, put it in a constant temperature water bath and let it stand , incubate at 65°C for 2 hours, during which time, mix up and down several times every 15-20min until clear, centrifuge at room temperature at 6000rpm for 10min, and transfer the supernatant to a new centrifuge tube. Add an equal volume of chloroform-isoamyl alcohol (24:1 v / v) for extraction twice, transfer the supernatant to a new centrifuge tube, add 0.6 times the volume of isopropanol, mix well, and let stand at room temperature for 1...
Embodiment 2
[0021] Example 2: Cloning process and sequencing
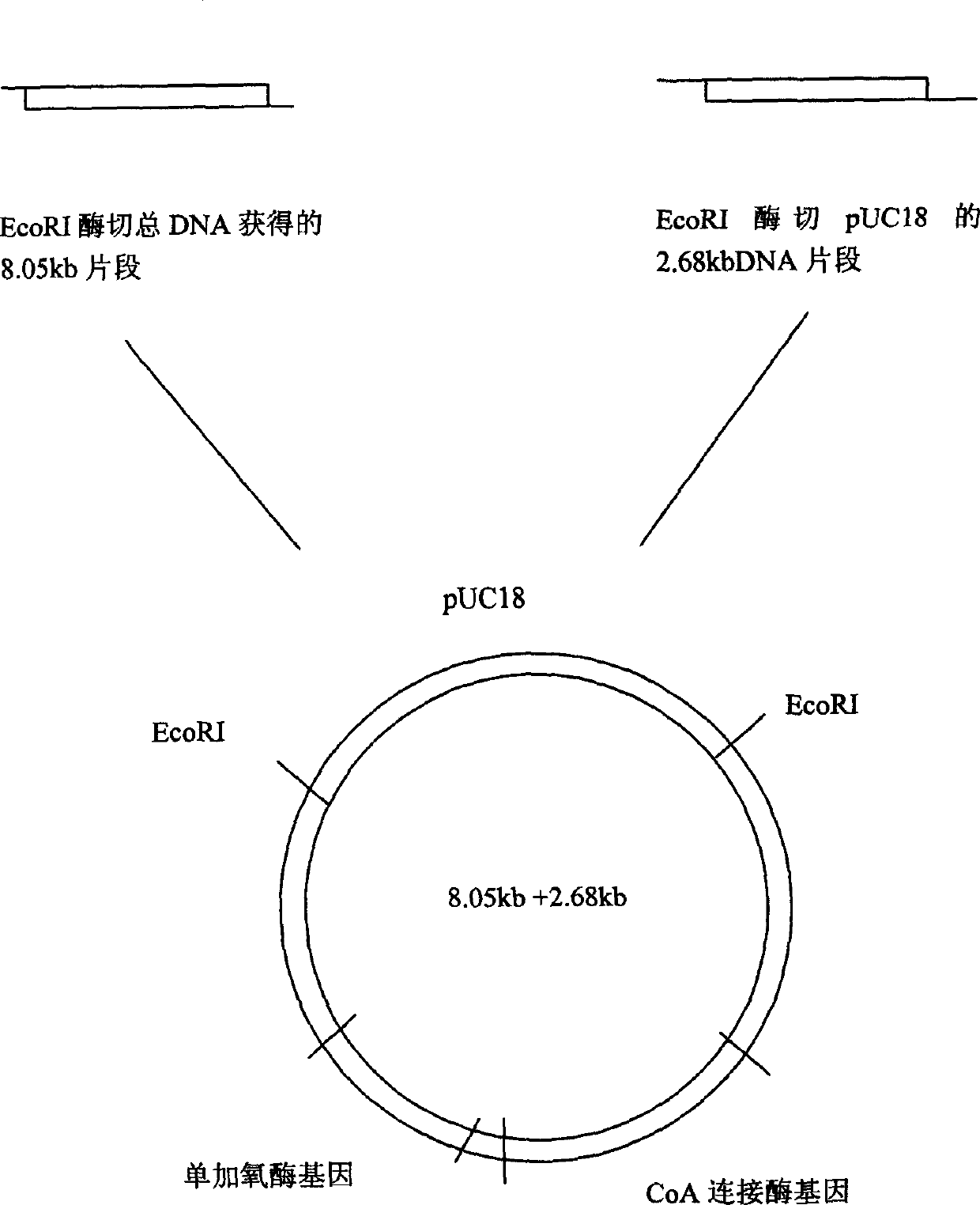
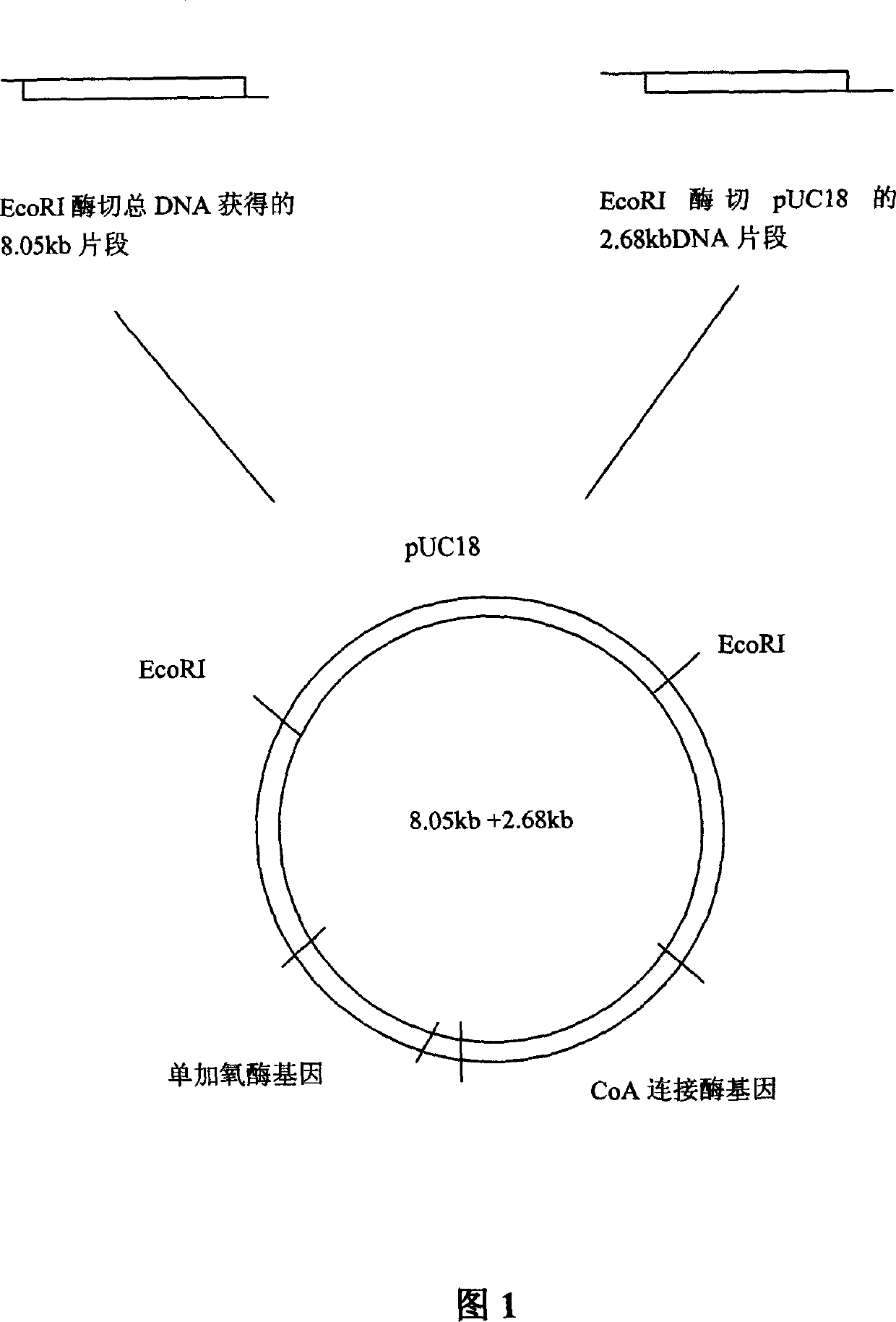
[0022] Take 10 μl of the above-mentioned total DNA solution, partially hydrolyze it with the restriction endonuclease EcoR I, and perform electrophoresis on agarose gel to obtain the digested fragments. Take 6 μl (5 μg) of enzymatically digested DNA fragments and 3 μl (2 μg) of EcoRI-digested pUC18 plasmid DNA, add 9ml TaKaRa DNA Ligation Kit Ver2.0 solution I to construct a ligation reaction system, and react at 16°C for 30 min; Competent cells of Escherichia coli HB101 prepared by calcium addition method. Transformants were plated on LB solid medium containing 75 μg / ml Amp (ampicillin). After culturing, several blue colonies grew on the transformant plate, and a recombinant strain capable of accumulating indigo in the transformant cells of Escherichia coli was obtained. Pick one of the single colonies for culture, extract the plasmid by alkaline lysis, hydrolyze the recombinant plasmid with restriction endonuclease, and co...
Embodiment 3
[0024] Embodiment 3: Verification test
[0025] (1) The oxygenation activity of the gene product was verified by the indigo generation experiment: indigo was dissolved in dimethylformamide (DMF), and at OD 610 There are specific absorption peaks. Observe OD 610 The amount of indigo produced can be calculated by the increase of light absorption value. The indigo molar absorptivity coefficient of the ultraviolet spectrophotometer is 17200 liters / mol.cm. In the M9 liquid medium containing 0.2% glucose (Amp 100mg / L), add indole 60umol / L as the substrate, and use the recombinant Escherichia coli to do the indigo production experiment. Take a certain amount of culture solution every 30 minutes, dissolve the indigo produced by oxygenase oxindole in DMF, and measure the OD 610 The absorbance value at the place can be used to measure the activity of oxygenase in recombinant cells. Experiments proved that the recombinant strains containing the above genes completely transformed mos...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com