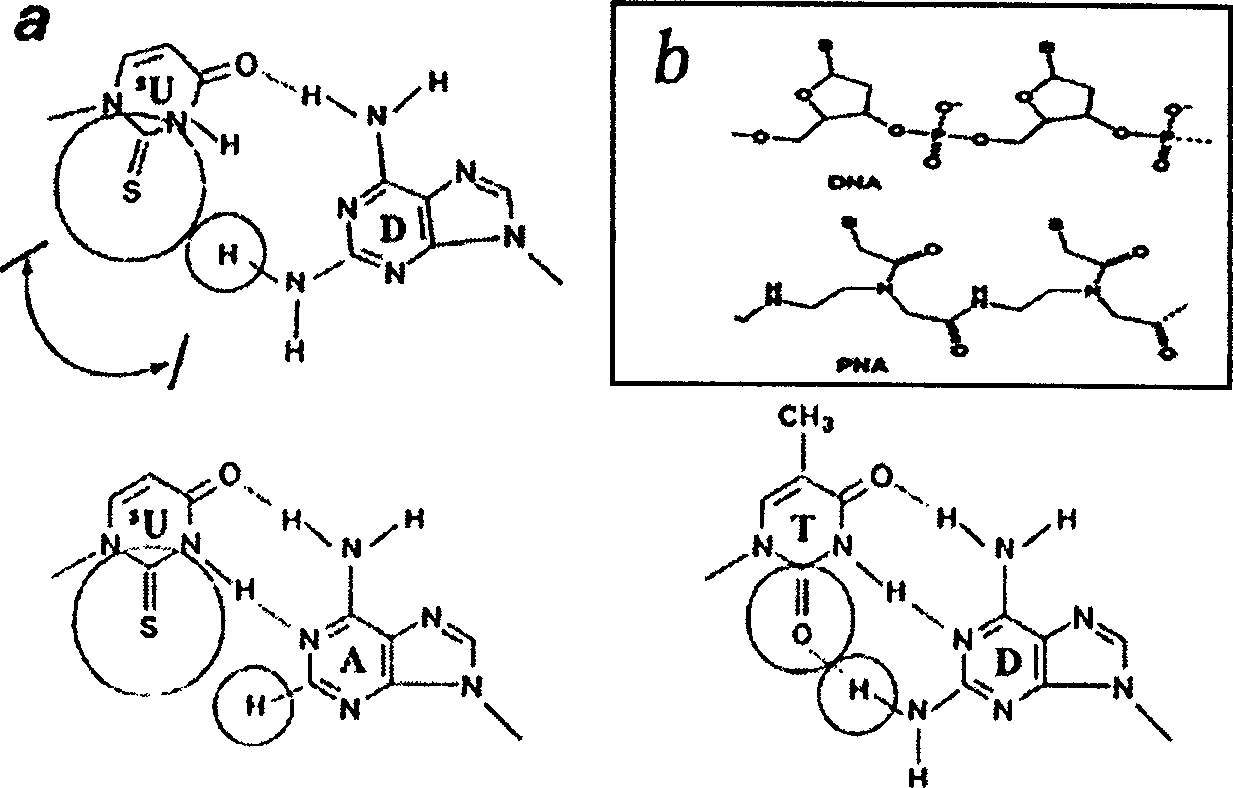
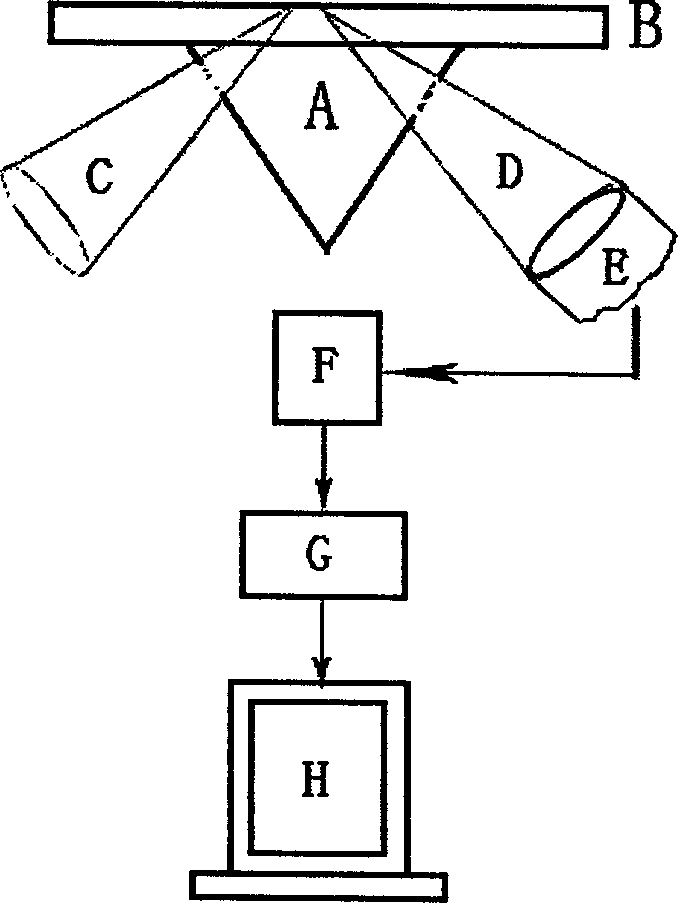
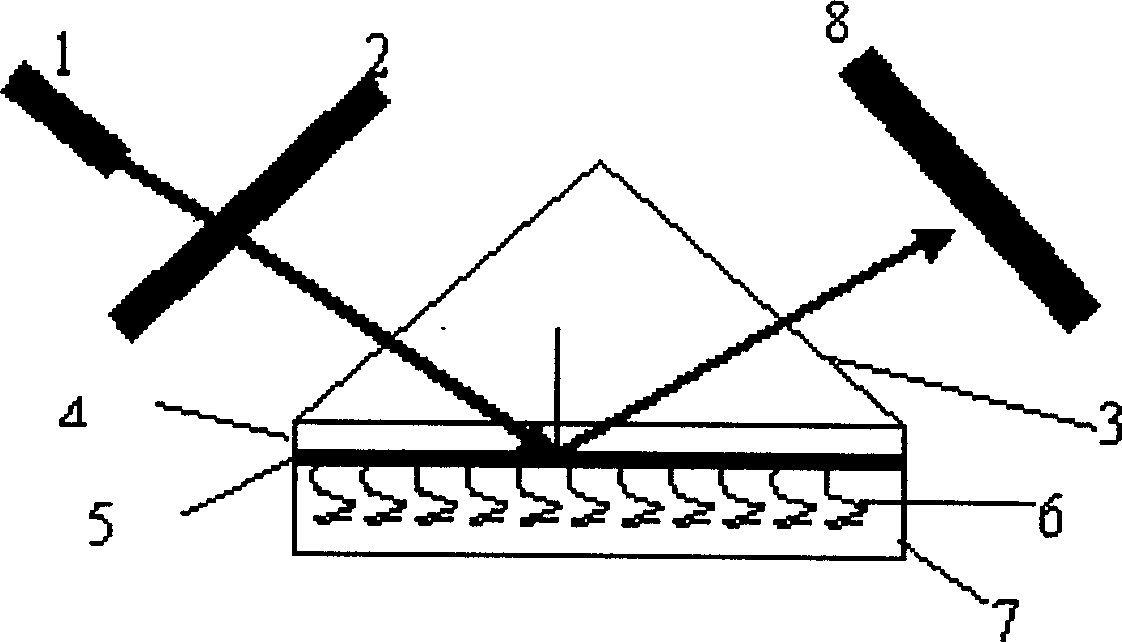
False complementary peptide nucleic acid probe biochip and detection method based on SPR principle
A biochip, peptide nucleic acid technology, applied in the determination/inspection of microorganisms, biochemical equipment and methods, etc., can solve the problems of reducing the universality and selectivity of PNA, reduce the cost of use and experimental conditions, and accurately detect and identify , high sensitivity and specificity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0055] Example 1 On a glass slide, a nano-gold film method with a surface plasmon resonance sensitivity of 50nm thickness is paved, and the steps are as follows:
[0056] (1), prepare gold sol by trisodium citrate reduction method (Frens method): get 100ml of 0.01% chloroauric acid aqueous solution and heat to boil, accurately add 0.75ml of 1% trisodium citrate aqueous solution under stirring, golden yellow chloroauric acid The aqueous solution turned purple within 2 minutes, continued to boil for 15 minutes, and returned to the original volume with distilled water after cooling.
[0057] (2), gold film paving:
[0058] A. After the slides are acid-washed and cleaned, put them in N-β-(aminoethyl)-γaminopropyl trivalent oxysilane (APTMS) / toluene solution and reflux for 12 hours, then wash them, take them out, and dry them with nitrogen , put in a clean and dry place for later use;
[0059] B. After soaking the treated glass slide in the nano-gold solution for 12 hours, immers...
Embodiment 2
[0060] Embodiment 2 The method of applying surface self-assembled monolayer technology to make pcPNAs probe array on the surface of gold film, the steps are as follows:
[0061] (1), prepared nano-gold film in piranha solution (30%H 2 o 2 : Concentrated H 2 SO 4 = 1: 3), then rinsed repeatedly with double distilled water, and dried with nitrogen;
[0062] (2), drop the SH-pcPNAs / PBS buffer solution of 500nM concentration and the self-assembly reaction on the nano-gold film, and react for 24 hours at 100% humidity to generate a self-assembled array of probes;
[0063] (3) Blot the buffer solution to dryness respectively, block with 6-mercaptohexanol, wash with double distilled water, and blow dry with nitrogen gas.
Embodiment 3
[0064] Example 3 One-step rapid processing of specimens with reagent Chelex-100 to extract bacterial DNA, the steps are as follows:
[0065] (1), collect one ring of pure culture colonies with a sterile inoculation loop, and place in a 1.5ml sterile Eppendorf tube;
[0066] (2) Add 50 μl of 5% Chelex suspension, mix well, add 20 mg / ml proteinase K, digest in a water bath at 56°C for more than 2 hours, then put it in a boiling water bath for 7.5 minutes, then immediately put it in an ice bath for 3 minutes, and centrifuge at 12000 r / min for 5 minutes , take the supernatant as a DNA sample.
PUM
| Property | Measurement | Unit |
|---|---|---|
| thickness | aaaaa | aaaaa |
| diameter | aaaaa | aaaaa |
| thickness | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com