Modulation of exon recognition in pre-mRNA by interfering with the secondary RNA structure
a technology of exon recognition and secondary rna, which is applied in the field of molecular biology and medicine, can solve the problems of reducing the risk that also one or more other pre-mrna will be able to hybridize to the oligonucleotide, disrupting the exon inclusion signal, etc., and achieves strong secondary structures. , the effect of improving the invasion efficiency of the oligonucleotide and increasing efficiency
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
Results
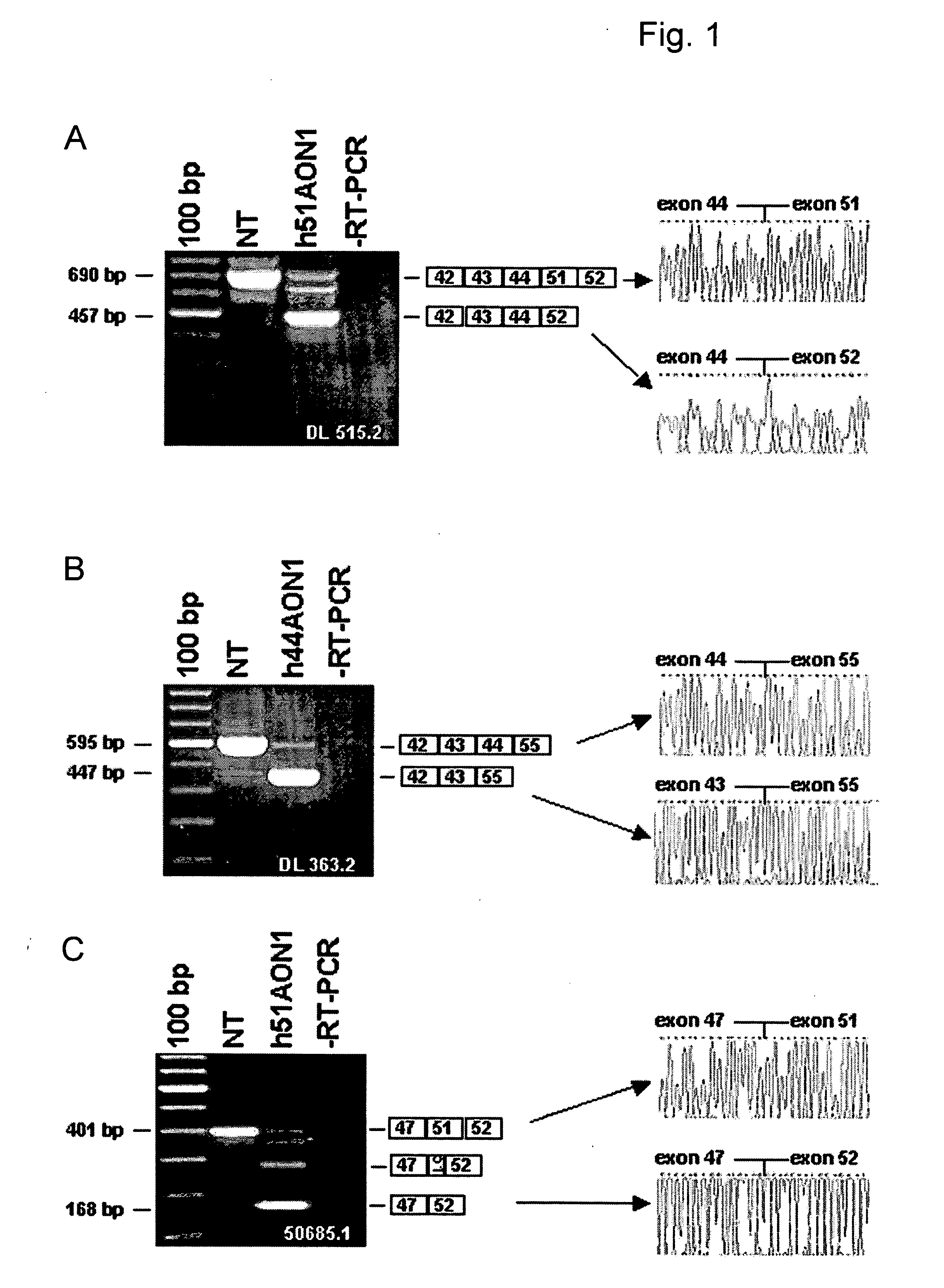
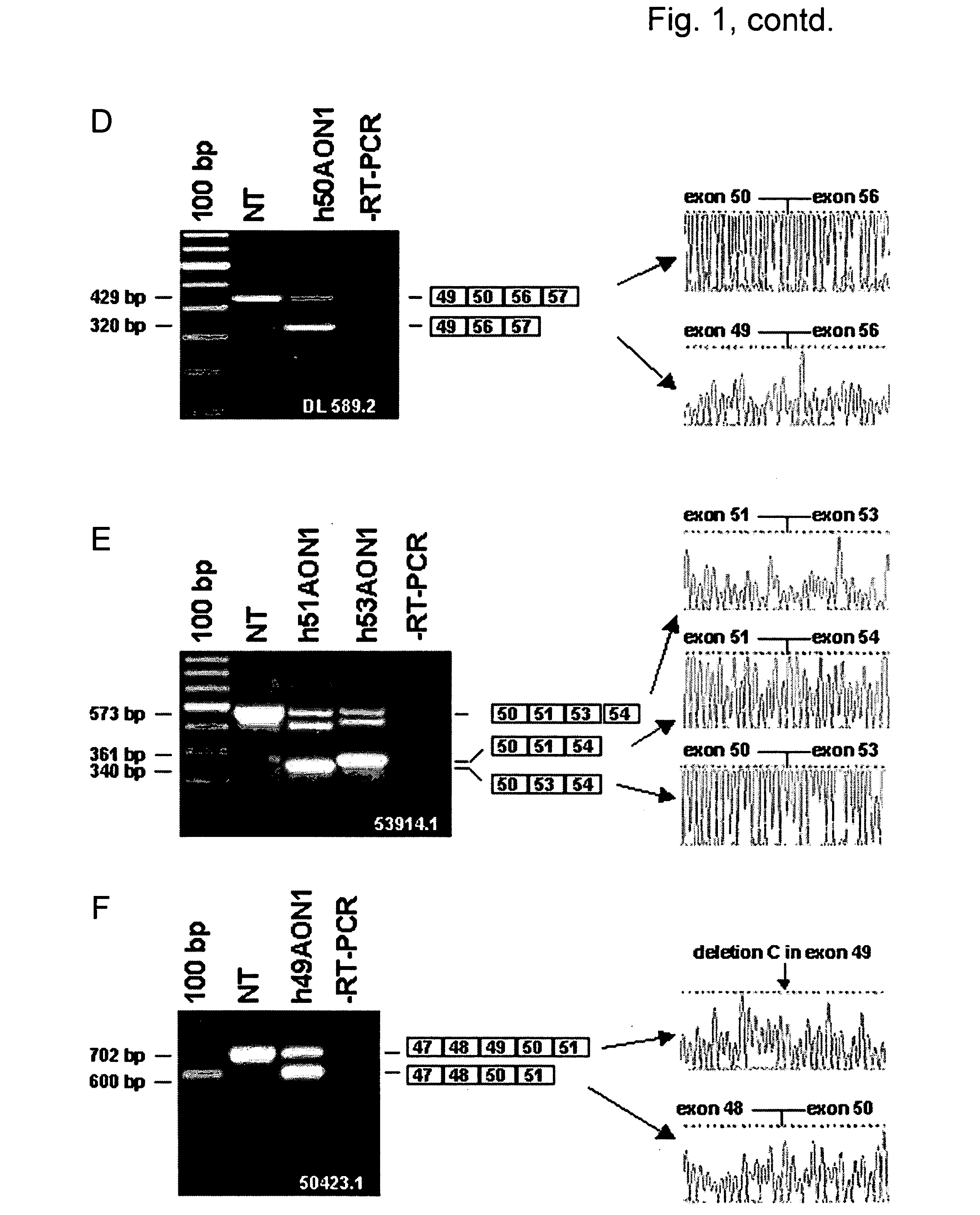
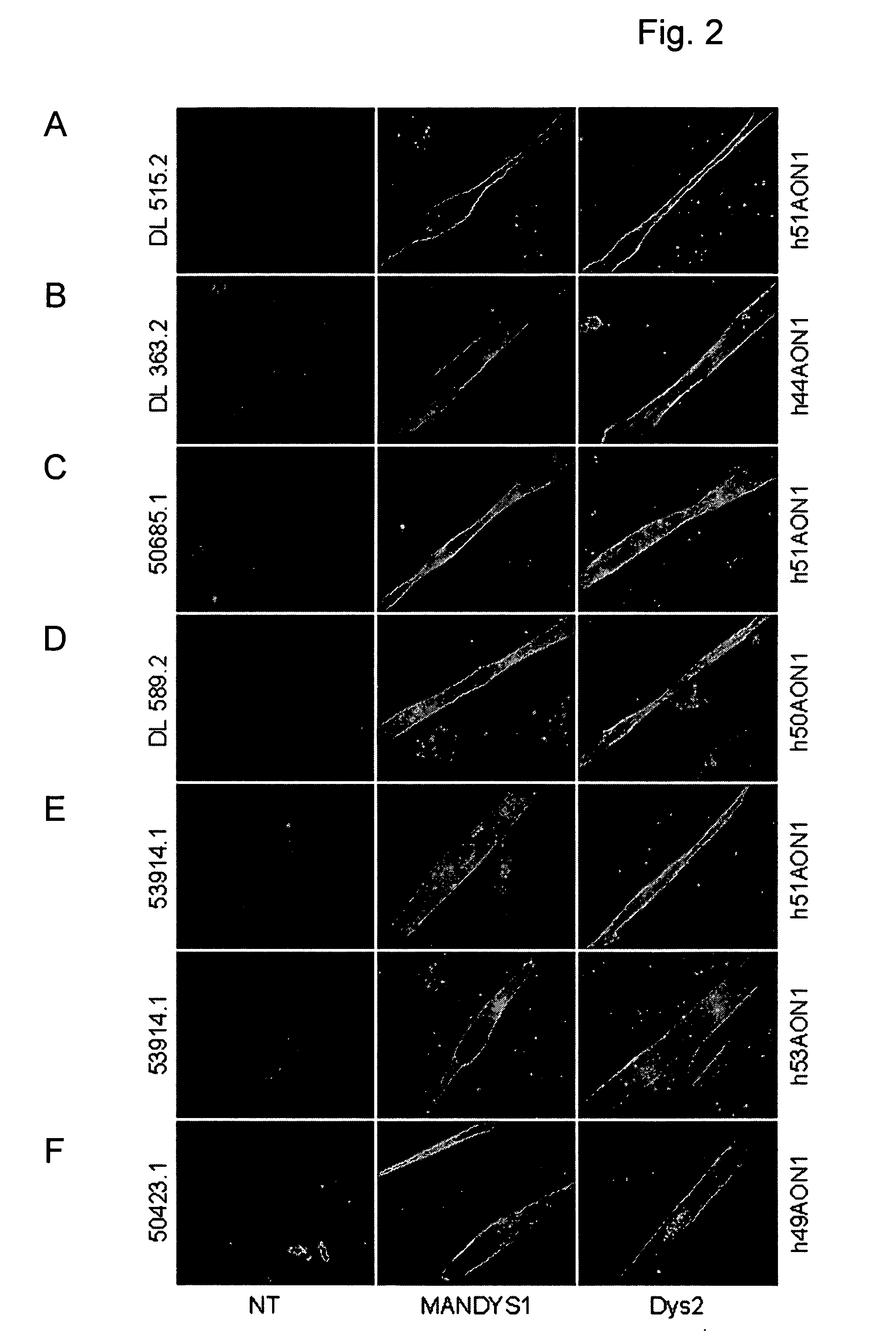
[0047] This study includes 6 DMD patients affected by different mutations (Table 1). Patient DL 515.2 carries an exon 45-50 deletion; hence exon 51 skipping would be frame correcting. Patient DL 363.2 has a deletion of exon 45-54; the reading frame for this patient would be corrected by an exon 44 skip. For patient 50685.1, who is affected by an exon 48-50 deletion, reading frame correction requires an exon 51 skip. Patient DL 589.2 has an exon 51-55 deletion; the reading frame would be corrected by an exon 50 skip. Patient 53914.1 carries a single exon 52 deletion. Notably, in this case both the skipping of exon 51 or exon 53 would be frame correcting. Finally, patient 50423.1 has a deletion of a single base pair in exon 49, at position 7389 on cDNA level, resulting in a frame-shift and a premature stop codon in exon 49. Since exon 49 is an in-frame exon, skipping of this exon would correct the reading frame for this patient.
[0048] We have previously identified AONs with ...
example 2
Materials and Methods
AONs and Primers
[0065] A series of AONs (two per exon, see Table 2) was designed to bind to exon-internal target sequences showing a relatively high purine-content and, preferably, an open secondary pre-mRNA structure (at 37° C.), as predicted by the RNA mfold version 3.1 server [22]. The AONs varied in length between 15 and 24 bp, with G / C contents between 26 and 67%. They were synthesized with the following chemical modifications: a 5′-fluorescein group (6-FAM), a full-length phosphorothioate backbone and 2′-O-methyl modified ribose molecules (Eurogentec, Belgium). The primers used for reverse transcription-polymerase chain reaction (RT-PCR) analysis (Table 3) were synthesized by Eurogentec (Belgium) or by Isogen Bioscience BV (The Netherlands).
In Vitro Experiments
[0066] Primary human myoblasts were isolated from a muscle biopsy from a non-affected individual (KM108) by enzymatic dissociation. Briefly, the tissue was homogenized in a solution containing...
example 3
Results
[0101] Double-Exon Skipping in Two DMD Patients
[0102] This study includes two DMD patients affected by different frame-disrupting mutations in the DMD gene that require the skip of two exons for correction of the reading frame (Table 5). Patient DL 90.3 carries a nonsense mutation in exon 43. Considering that this single exon is out-of-frame, the skipping of exon 43 would remove the nonsense mutation but not restore the reading frame. Since the combination with exon 44 is in-frame, we aimed in this patient at double-exon skipping, targeting both these exons. Patient DL 470.2 is affected by a deletion of exons 46 to 50. Frame restoration would require a double-exon skipping of both exons flanking the deletion. Myotube cultures from both patients were transfected with a mixture of exon 43 and 44 specific AONs (DL90.3) or exon 45 and 51 specific AONs (DL470.2). The individual AONs (Table 5) were previously highly effective in single exon skipping. Transfection efficiencies we...
PUM
| Property | Measurement | Unit |
|---|---|---|
| pH | aaaaa | aaaaa |
| pH | aaaaa | aaaaa |
| pH | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com