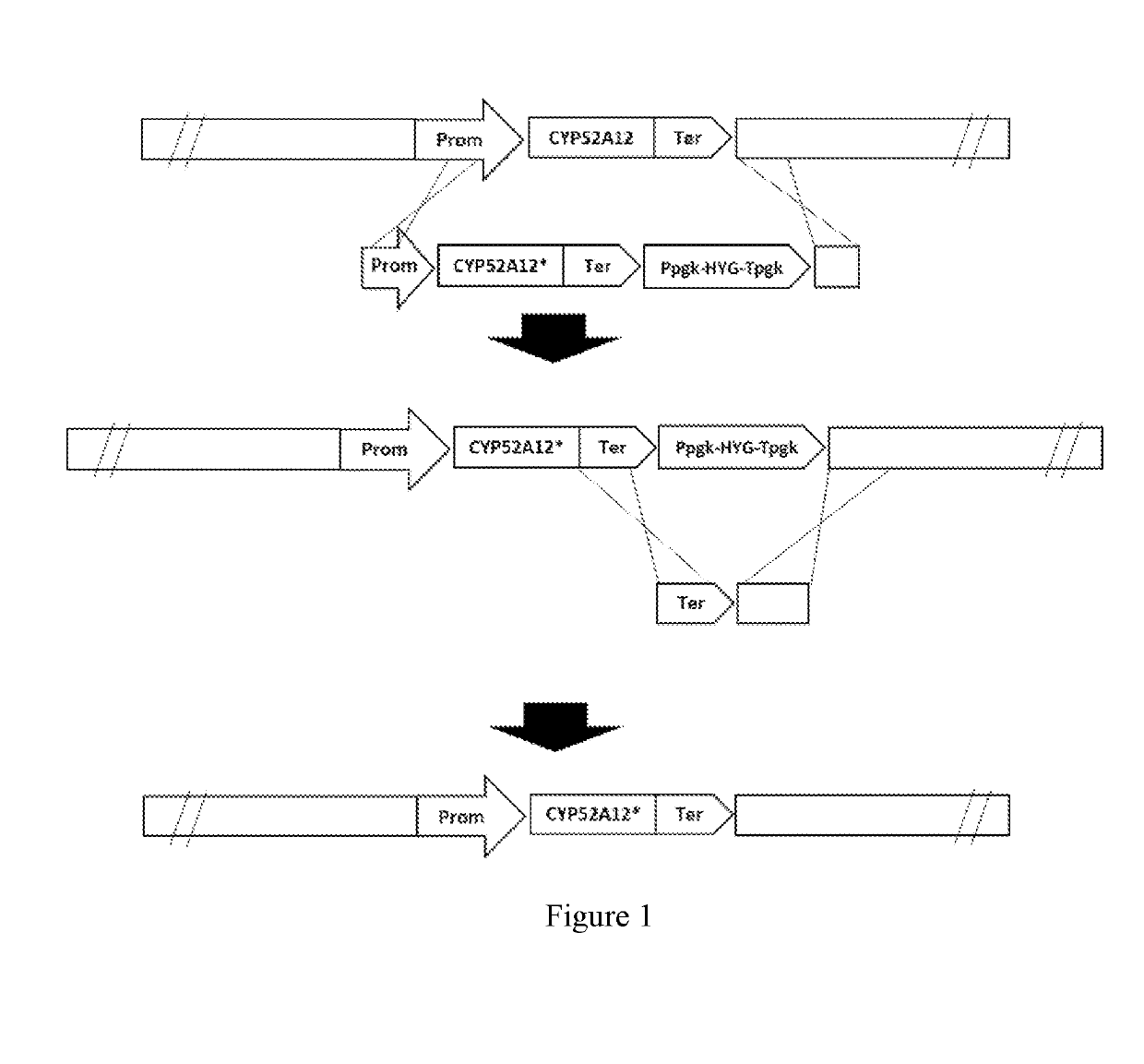
Directed evolution of cyp52a12 gene and its use in dicarboxylic acid production
a technology of cyp52a12 and dicarboxylic acid, which is applied in the field of preparing a long-chain dicarboxylic acid producing strain, can solve the problems of huge obstacles to serious affecting the development of long-chain dicarboxylic acid in the industry, and many challenges in chemical synthesis. achieve the effect of significantly shortening the fermentation cycl
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
edia and Methods for Culture and Fermentation as Well as for Detecting a Dicarboxylic Acid
[0086]1. YPD medium formula (w / v) was: 2% peptone, 2% glucose and 1% yeast extract (OXOID, LP0021). 1.5-2% agar powder was added to form a solid media.
[0087]During culturing, a single colony was picked in a 2 ml centrifuge tube containing 1 ml YPD liquid medium, incubated at 30° C. in a 250 RPM shaker for 1 day.
[0088]2. Seed medium formula (w / v): sucrose 10 to 20 g / L, yeast extract 3 to 8 g / L, industrial fermentation corn syrup (for short, corn syrup, with total nitrogen content 2.5 wt %) 2 to 4 g / L, KH2PO4 4 to 12 g / L, urea 0.5 to 4 g / L (separately sterilized at 115° C. for 20 min), and the substrate for fermentation was n-dodecane 20 mL / L.
[0089]During culturing, the inoculum obtained in step 1 was inoculated into a 500 mL shake flask containing 30 mL seed medium, wherein the amount of inoculum was 3-5%, and incubated at 30° C. in a 250 RPM shaker until OD620 reached 0.8 (after 30-fold dilutio...
example 2
on of CYP52A12 Mutation Template
[0094]1. Preparation of the CYP52A12 mutation template.
[0095]The genomic DNA of Candida CCTCC M 2011192 was extracted by using Ezup Yeast Genomic DNA Extraction Kit (Sangon, Cat No. 518257). A method with liquid nitrogen grinding was used in favor of increasing the cell wall disruption efficiency. Genomic DNA obtained by this method was used as template for error-prone PCR.
[0096]2. Error-prone PCR
[0097]The concentration of Mg2+ was adjusted (2-8 mM) and the CYP52A12 gene was amplified by error-prone PCR using Taq DNA Polymerase (Takara, Cat No. R00113).
[0098](PCR condition was: Step 1: 98° C. for 30 s, step 2: 98° C. for 10 s, 55° C. for 30 s, 72° C. for 2 m 20 s, 35 cycles in total, Step 3: 72° C. for 5 m).
[0099]The primers were as follows:
CYP52A12-F:(SEQ ID NO: 1)5′-CAAAACAGCACTCCGCTTGT-3′,CYP52A12-R:(SEQ ID NO: 2)5′-GGATGACGTGTGTGGCTTGA-3′,
[0100]The PCR product was subjected to electrophoresis on a 1% agarose gel and recovered and purified by using...
example 3
on of Homologous Recombination Template
[0101]All DNA fragments in this example were obtained by amplification using PrimeSTAR® HS High Fidelity DNA polymerase (Takara, R040A). The DNA fragments were subjected to electrophoresis on a 1% agarose gel, followed by recovery and purification by using the Axygen Gel Recovery Kit.
[0102](1) Amplification of the resistance selection marker (HYG, the hygromycin resistance gene). The amplification template was the vector pCIB2 (SEQ ID NO: 3) owned by our company. The primer sequences were as follows:
CYP52A12_HYG-F:(SEQ ID NO: 4)5′-TCAAGCCACACACGTCATCCGCATGCGAACCCGAAAATGG-3′,CYP52A12_HYG-R:(SEQ ID NO: 5)5′-GATGTGGTGATGGGTGGGCTGCTAGCAGCTGGATTTCACT-3′.
[0103]The PCR reaction condition was as follows:
[0104]Step 1: 98° C. for 30 s,
[0105]Step 2: 98° C. for 10 s, 55° C. for 30 s, 72° C. for 1 m 50 s, 5 cycles,
[0106]Step 3: 98° C. for 10 s, 72° C. for 2 m, 25 cycles,
[0107]Step 4: 72° C. for 5 m.
[0108]The resulting product, named HYG, was verified by seq...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com