Using mutations to improve Aspergillus phytases
a technology of aspergillus and mutations, applied in the field of using mutations to improve the phytase of aspergillus, can solve the problems of false activation, low specific activity of this phytase, and no studies have successfully employed any information to improve this widely used benchmark phytase, etc., to improve the nutritional value of a food consumed by humans, increase the bioavailability of minerals, and improve the nutritional value of a food
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
Using Mutations to Improve Aspergillus Phytases
[0117]As used in Examples 1-13, amino acid residue 27 of the Aspergillus niger phytase corresponds to amino acid residue 50 as referenced in SEQ ID NOS:2, 6, and 12, and in the claims of the present application. Also, as used in Examples 1-13, amino acid residue 362 of the Aspergillus fumigatus phytase corresponds to amino acid residue 363 as referenced in SEQ ID NOS:4 and 10 and in the claims of the present application.
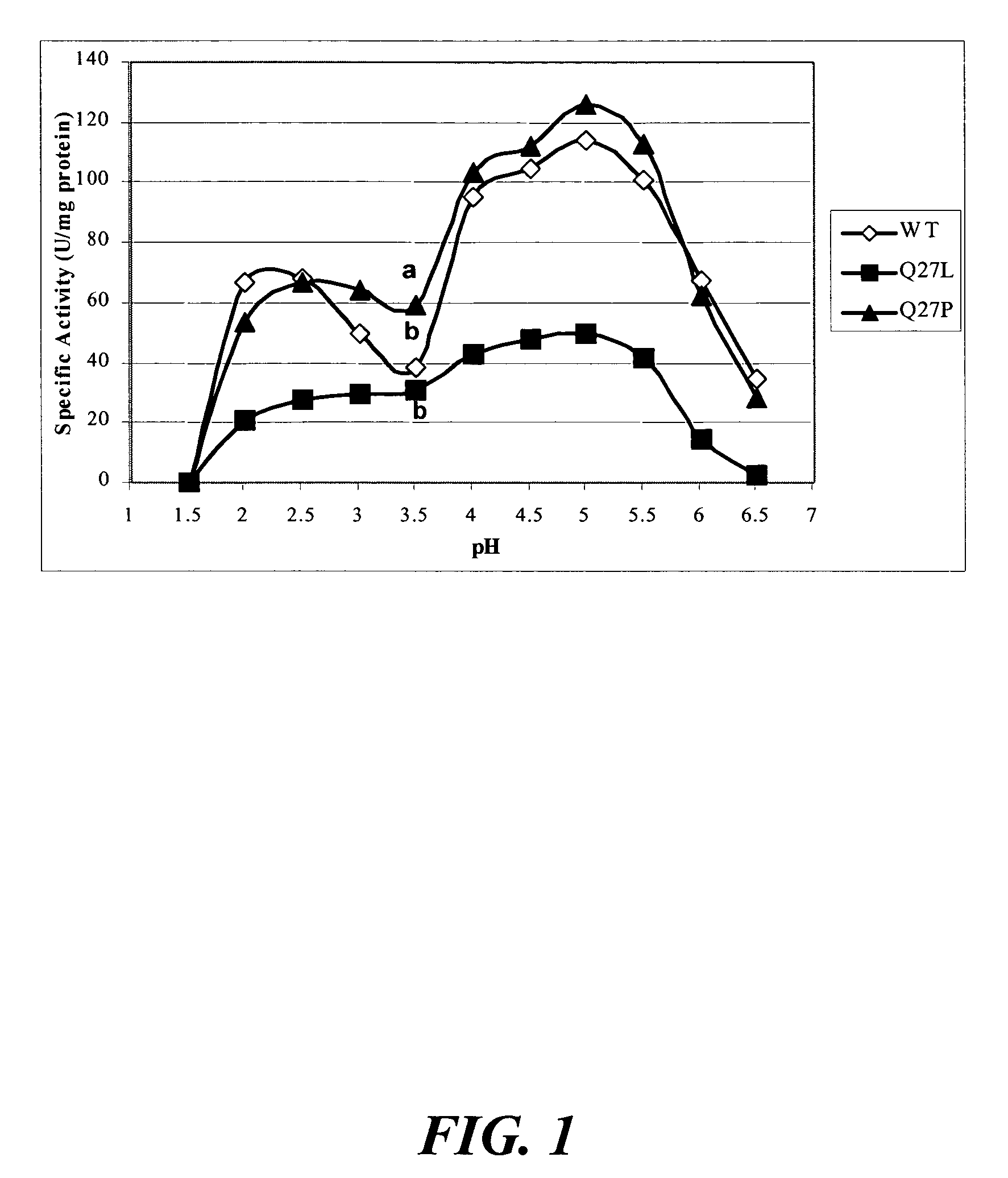
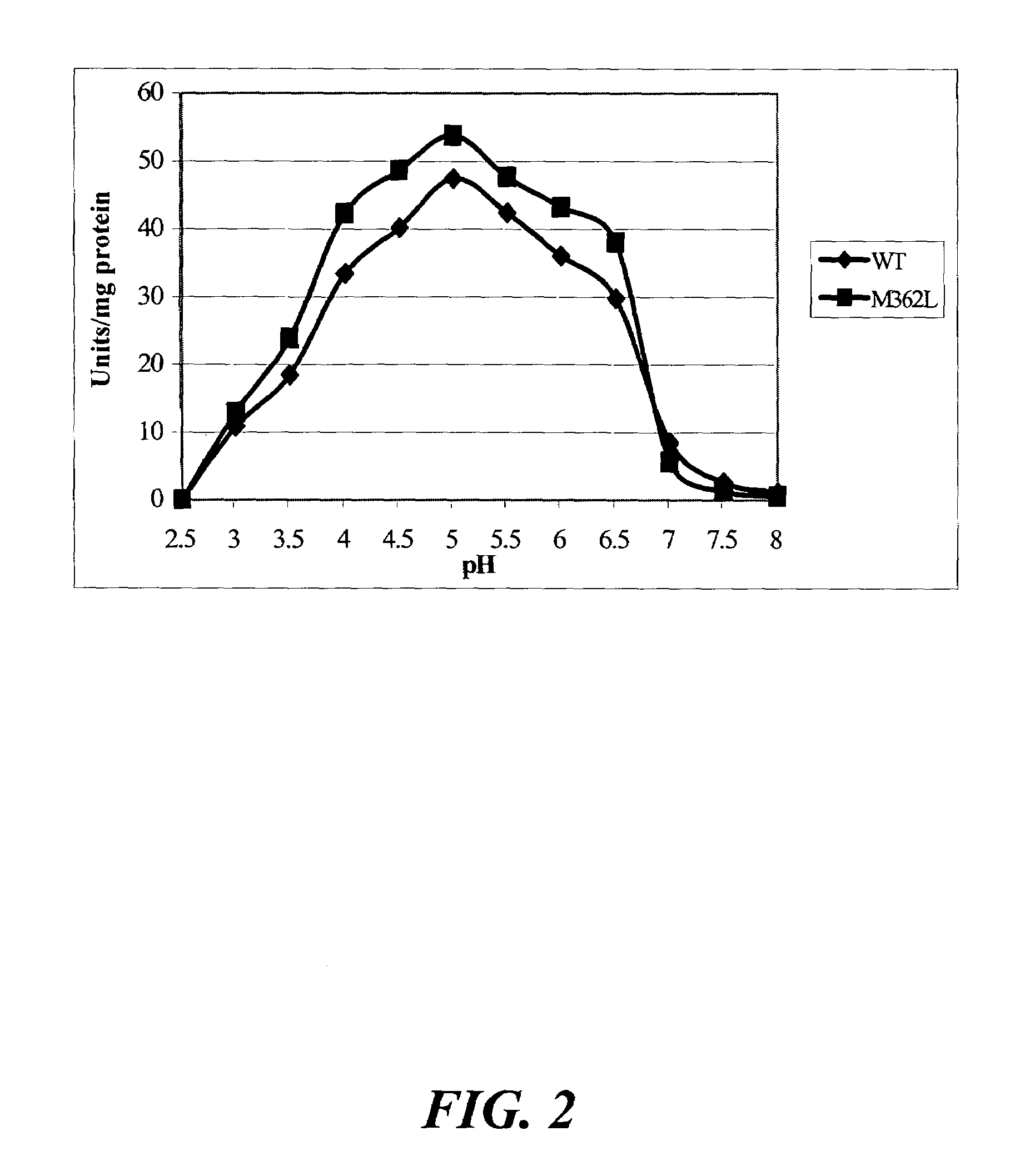
[0118]The objectives of this study included the following: (1) to compare the specific activity of mutants Q27L and Q27P in A. niger phytase as well as M362L in A. fumigatus phytase with the respective wild-type controls; and (2) to determine the impacts of these single amino acid substitutions on the pH profile and heat-tolerance of the recombinant phytases.
[0119]Site-directed mutagenesis was conducted to enhance catalytic activities of Aspergillus niger and A. fumigatus phytases. Mutation Q27L in A. niger phytase cause...
example 2
Phytase Mutations
[0120]Plasmid pYPP1 containing the cloned A. niger NRRL 3135 phyA phytase gene (Han et al., “Expression of an Aspergillus niger Phytase Gene (phyA) in Saccharomyces cerevisiae,” Appl. Environ. Microbiol. 65:1915-1918 (1999), which is hereby incorporated by reference in its entirety) was utilized to generate two mutants of Q27L and Q27P. Based on the published sequence of this phyA phytase gene (GeneBank accession no. M94550), the following oligonucleotides were synthesized to generate site specific mutations at the Gln 27 residue, corresponding to the same residue in A. fumigatus (Tomschy et al., “Optimization of the Catalytic Properties of Aspergillus fumigatus Phytase Based on the Three-Dimensional Structure,”Protein Science 9:1304-1311 (2000), which is hereby incorporated by reference in its entirety), Leu 27 5′-CTTTGGGGTCTATACGCACCG-3′ (SEQ ID NO:57) and Pro 27 5′-CTTTGGGGTCCATACGCACCG-3′ (SEQ ID NO:58). The primers were phosphorylated and the Gene Editor™ in vi...
example 3
Yeast Transformation and Protein Expression
[0122]Saccharomyces cerevisiae INVSc1 (Invitrogen) were grown in yeast extract-peptone-dextrose medium (YPD) and prepared for transformation according to the manufacturer instructions. Plasmid DNA containing Q27L or Q27P was transformed into Saccharomyces by electroporation (1.5 KV, 50 μF, 129 Ω. ECM 600 Electro Cell Manipulator, Genetronics, BTX Instrument Division, San Diego, Calif.). After incubation for 2 h at 30° C. in 1 M sorbitol without agitation, cells were plated in URA(−) selective medium to screen for positive transformants. Colonies were grown in 9 mL YPD broth for 36-48 h, and then centrifuged at 1,500 rpm, 25° C. for 10 min. The cell pellet was resuspended in YPG medium (1% Yeast extract, 2% peptone, 2% galactose) for induction of the recombinant enzyme expression. Activity in the medium was measured after 24-36 h. Transformation of plasmid DNA containing M362L in Pichia pastoris strain X33, and induction of phytase expressio...
PUM
| Property | Measurement | Unit |
|---|---|---|
| pH | aaaaa | aaaaa |
| pH | aaaaa | aaaaa |
| pH | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com