Gene chip and kit for detecting invasive fungal pathogenic bacteria
An invasive fungal, gene chip technology, applied in the directions of microorganism-based methods, biochemical equipment and methods, and microbial determination/examination, which can solve the problems of difficulty in diagnosing fungal infection, difficulty in samples, false positives, etc., and shorten the detection time. cycle, improving detection sensitivity and specificity, and the effect of high accuracy
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0035] Design and preparation of embodiment 1 probe
[0036]Ribosomal DNA sequences exist in all organisms, have the same origin and function during evolution, and can reflect comparable evolutionary histories among species. The internal transcribed spacer (Internal Transcribed Spacer, ITS) of rDNA is a region between the 18S and 28S rDNA gene copy regions in eukaryotic rDNA, and the 5.8S rDNA is located between ITS1 and ITS2. The nucleotide sequence of 5.8SrDNA is highly conserved, while ITS1 and ITS2 evolve rapidly, but the inter-species homology is very small, while the intra-species homology is very high, which can be used for inter-species identification.
[0037] 1. Sequence acquisition:
[0038] (1) Acquisition of the 18S-28S region gene sequence: Candida albicans, Candida parapsilosis, Candida tropicalis, Candida kefir, Candida krusei, and Pseudomonas portuguese were downloaded from the GenBank public database. The 18S-28S region gene sequences of Trichomonas, Cand...
Embodiment 2
[0051] Design and preparation of embodiment 2 universal primers
[0052] 1. Universal primer sequence:
[0053]
[0054] 2. Primer synthesis: Entrust the primer synthesis company (Beijing Handsome) to synthesize the primer sequences in the above table, and reserve them for later use.
Embodiment 3
[0055] Example 3 Gene Chip Preparation——Chip Spotting
[0056] 1. Dissolving probes: the probes synthesized in Example 1 were respectively dissolved in 50% DMSO solution, and diluted so that the final concentration of the probes reached 1 μg / μl.
[0057] 2. Adding plate: Add the dissolved probe to the corresponding position of the 384-well plate, 10 μl per well.
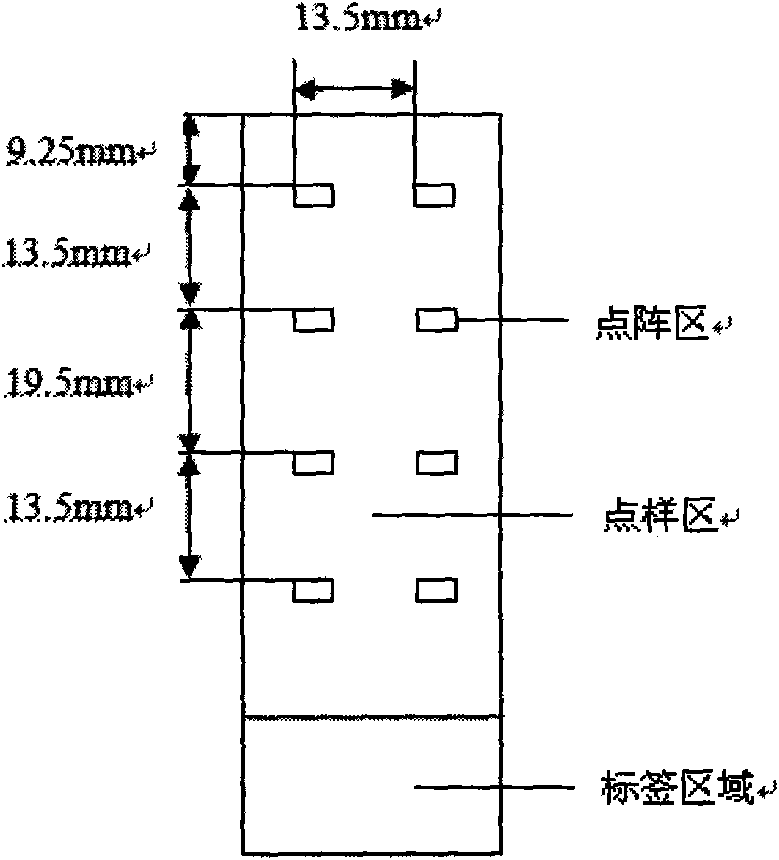
[0058] 3. Spotting: as figure 1 The shown 75.5mm×25.5mm×1mm (length×width×height) clean aldehydated glass slide is placed on the stage of the chip spotting instrument (SpotArray 72), and the control software of SpotArray is used to run the program. according to figure 2 The arrangement shown is spotted on the aldehyde-formed glass slide in the spotting area of 4.5 mm × 4.5 mm to form a medium-low density DNA micro-array, and the eight dot matrix areas on the glass slide have the same array arrangement rule. The size of the dot matrix area is 3.75mm×2mm, the dot pitch in the dot matrix is 250μm, the matrix: ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com