Screening and application of probiotic Enterococcus faecium
A technology of Enterococcus faecium and Staphylococcus faecium, applied in the direction of application, microbial-based methods, bacteria, etc., can solve problems such as endangering the healthy development of aquaculture, affecting human food safety, and imbalance of normal animal flora, so as to improve livestock and poultry production Effects on performance, human and animal safety, production and ease of use
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0046] Example 1: Isolation and identification of probiotic strains
[0047] 1. Isolation of strains
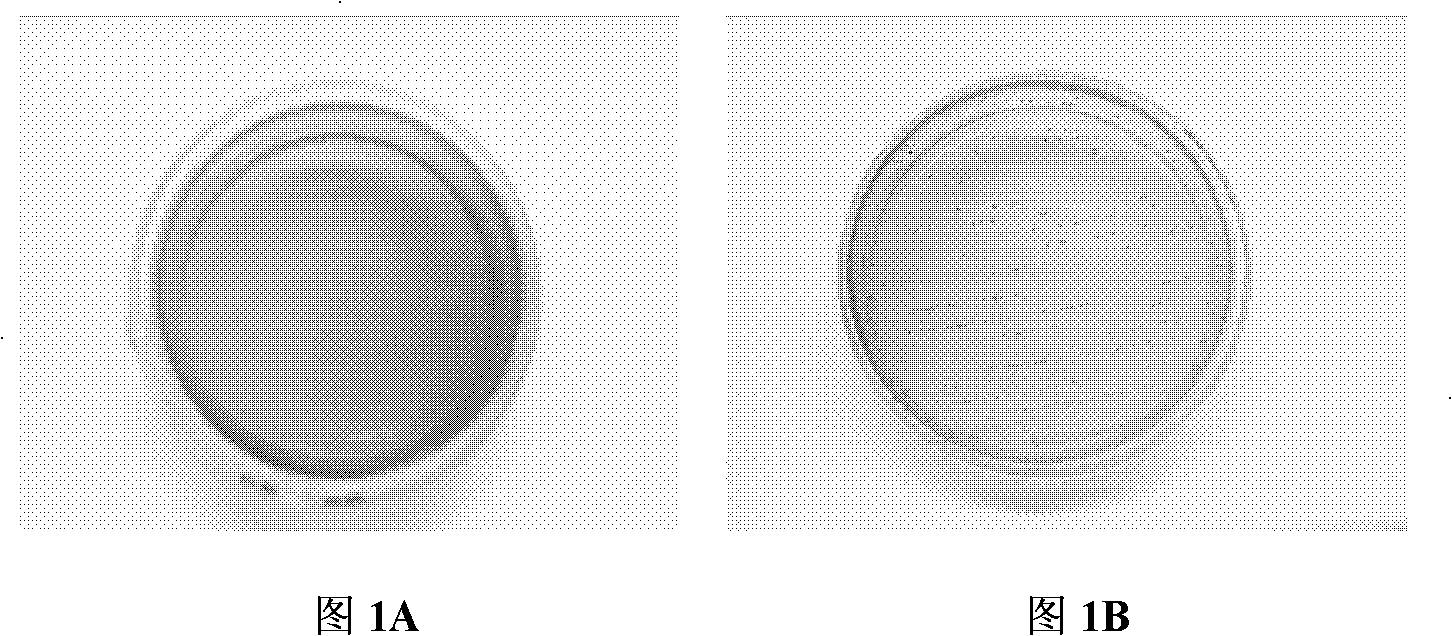
[0048] The samples were taken from the 100-day-old and 21-day-old Chinese local pig breed Hubei "Tongcheng Pig" (the breed comes from the breeding farm of Tongcheng County, Hubei Province), and the rectal contents of the pigs were collected from the anus with sterilized cotton swabs. After the sample is collected, use a cotton swab to streak and culture it in KF Streptococcus agar medium (purchased from Hangzhou Microbial Reagent Co., Ltd.). Colonies of large size were purely cultured. The pure culture was preliminarily screened by colony morphology observation, Gram staining and peroxidase test. Bacteria in the form of single, paired or short-chain oval Gram-positive cocci and strains negative for peroxidase tests are all passaged and preserved. The growth characteristic of the bacterial strain of this invention on the KF Streptococcus agar medium sees figure 1 , for its...
Embodiment 2
[0089] Embodiment 2: Bacteriostatic performance test of Enterococcus faecium strain HDRsEf1
[0090]Staphylococcus aureus (ATCC25923, purchased from the Clinical Inspection Center of the Ministry of Health), Salmonella choleraesuis C78-1 (purchased from China Veterinary Drug Administration) and porcine pathogenic Escherichia coli O138:K88 (C83902, purchased from Supervising Institute) is indicator bacterium, with HDRsEf1 bacterial strain fermentation supernatant of the present invention is antibacterial agent, detects the in vitro bacteriostatic performance of Enterococcus faecium bacterial strain HDRsEf1, concrete operation is as follows:
[0091] 1) Preparation and treatment of bacterial fermentation broth: Inoculate fresh Enterococcus faecium HDRsEf1 bacterial fluid into MRS broth medium (product of BD Company, USA) at 2% (v / v), culture it statically for 18-24 hours, and take it by centrifugation. The supernatant was centrifuged at 10000rpm for 30min, and the supernatant wa...
Embodiment 3
[0100] Example 3: Safety evaluation of Enterococcus faecium strains
[0101] 1. Amplification of virulence factors
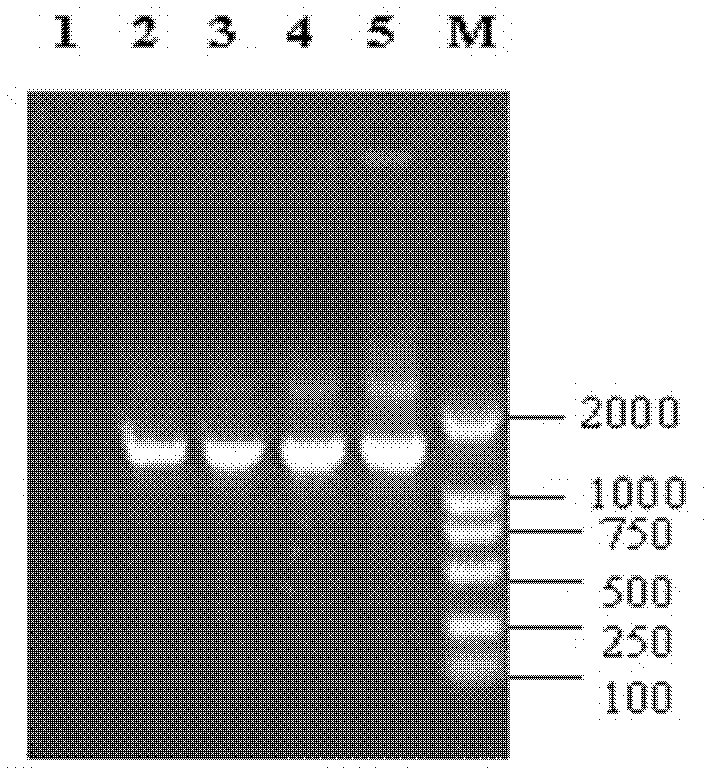
[0102] The amplification of virulence factors mainly includes cylA (accession number: AAA62652.1; Gilmore et al., 1994), gelE (accession number: D85392; Mannu et al., 2003), esp (accession number: AF034779; Shankar et al. ., 1999), agg (no accession number: Galli et al., 1990), ace (accession number: AF26083; Mannu et al., 2003), efaAfs (accession number: EFU03756) and efaA fm (Accession number: FJ609170.1; Mannu et al., 2003).
[0103] Using the Enterococcus genome prepared above as a template, PCR amplification was performed with specific primer pairs for each virulence factor. Each primer was synthesized by Shanghai Yingjun Biotechnology Co., Ltd., its sequence, PCR product size, amplification conditions and references See Table 4, and see Table 5 for the virulence gene PCR amplification system.
[0104] Table 4 Test primer sequences, PCR product size and ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com