Surface displaying system for rhizopus oryzaelipase, and preparation method and application of surface displaying system
A technology of rhizopus oryzae lipase and surface display system, which is applied in the field of genetic engineering, can solve the problems of long catalysis time, low yield of methyl esterification products, and easy shedding of displayed proteins, so as to reduce toxic and side effects, easy to operate, and good The effect of economic benefits and social effects
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0031] Example 1 S. cerevisiae The extraction and PCR amplification of the EBY100 genome yielded the mature ankyrin gene mpir1;
[0032] 1) Saccharomyces cerevisiae EBY100 ( S. cerevisiaeEBY100) culture: activate the strain on the YPD plate, pick a single colony in the YPD shake flask, and culture overnight at 30°C. Centrifuge at 6000rmp for 10min at 4°C to collect the strains, add 10% glycerol, and store the strains in a -80°C refrigerator. S. cerevisiae EBY100 was purchased from Invitrogen.
[0033] 2) Extraction of Saccharomyces cerevisiae EBY100 genomic DNA, steps: Pick a single colony from the activated Saccharomyces cerevisiae EBY100 plate, inoculate it in YPD medium, and culture it overnight at 30 ℃; take the cells in the logarithmic growth phase, centrifuge at 6000rpm for 5min, and the bacteria Wash the body once with sterile distilled water, remove the supernatant; add 5mL DNA wall breaking buffer (100mmol / L Tris-HCl, pH8.0, 10mmol / L EDTA, 1%SDS), mix well, an...
Embodiment 2
[0039] Example 2 Construction of surface display recombinant plasmids;
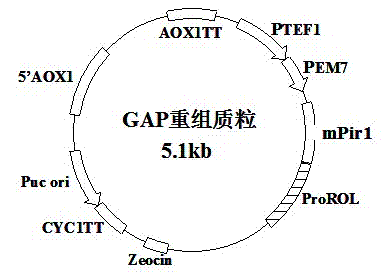
[0040] (1) Construction of AOX-mPir1 and GAP-mPir1 recombinant vectors
[0041] mPir1 (prepared in Example 1) and pPICZαA (Invitrogen) and pGAPZαA (Invitrogen) were digested with EcoRI and KpnⅠ, purified and recovered with the kit, and ligated with T4 DNA ligase to obtain the recombinant Pichia pastoris surface display expression plasmid GAP -mPir1, AOX-mPir1. The resulting GAP-mPir1 and AOX-mPir1 plasmids were transformed into Escherichia coli host TOP10F (Novagen). Transformants were screened with 50 μg / ml zeocin LLB plates, zeocin-positive transformants were picked, plasmids were extracted, identified by EcoRI and KpnⅠ double enzyme digestion and sequenced, the results showed that the wall protein mPir1p gene sequence was inserted correctly.
[0042] (2) Construction of AOX-mPir1-ProROL and GAP-mPir1-ProROL vectors
[0043] Cloning of the Rhizopus oryzae lipase gene prorol with a flag tag protein, f...
Embodiment 3
[0048] Example 3 Expression and identification of recombinant plasmids in Pichia pastoris;
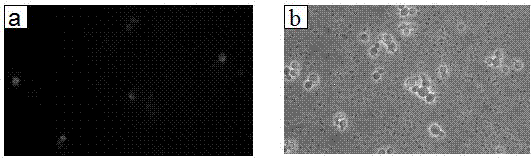
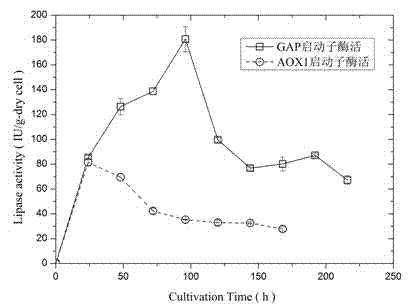
[0049] The AOX-mPir1-ProROL plasmid was linearized with restriction enzyme sac I, and the GAP-mPir1-ProROL plasmid was linearized with AvrII. Pichia pastoris KM71H (Invitrogen) was electrotransformed, grown on a YPD plate containing 100 μg / mL zeocin for 72 hours, and resistance-positive transformants were picked, that is, transformants integrated with fusion gene sequences (KM71H / AOX-mPir1- ProROL, KM71H / GAP-mPir1-ProROL), spot on the high-resistant YPD plate containing 500 μg / mL zeocin, select larger strains, that is, transformants containing more copies, and spot on the olive oil-MMH-RB plate to form The obvious fluorescent circle indicates that ProROL is expressed in Pichia pastoris and exhibits lipase hydrolysis activity.
[0050] Inoculate the positive transformants KM71H / AOX-mPir1-ProROL, KM71H / GAP-mPir1-ProROL in 50mL BMGY medium, culture at 30°C, 200rpm shaking for 16-20h to O...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com