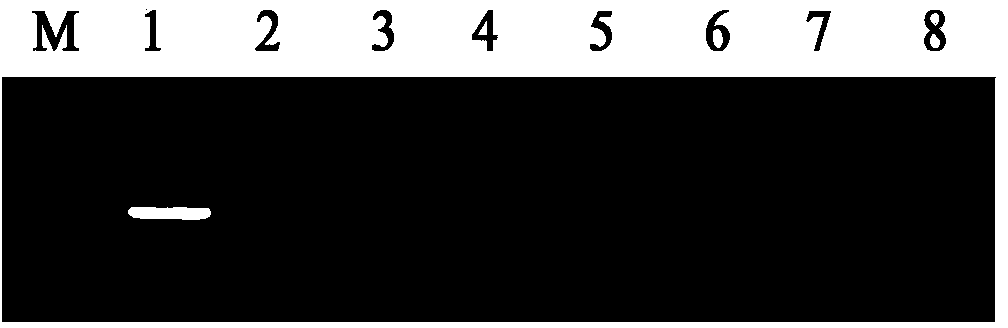Method for quantitatively detecting ustilaginoidea virens from seed-borne and soil-borne media
A technology for quantitative detection of rice false smut bacteria, applied in the field of microorganisms, can solve the problems of unsuitable for large-scale rapid application in the field, long detection time, high detection cost, etc., and achieve high specificity, simple method, and high sensitivity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0040] 1. Pre-treat the tested samples and quickly extract the DNA of the tested samples
[0041] (1) Seed DNA extraction steps: Weigh the rice seeds according to the thousand-grain weight standard, collect the seed shells after threshing, and operate according to the operating instructions of the Promega Plant Genomic DNA Extraction Kit (Promega (Beijing) Biotechnology Co., Ltd.) .
[0042] (2) Soil DNA extraction step: add 2ml SPCB buffer (100mmol / LTris-HCl, 100mmol / L EDTA, 120mM Na 3 PO 4 , mass fraction is 2% CTAB, 1.5M NaCl, pH8.0, volume percentage is 2% mercaptoethanol), the ratio of 100 μ g / ml proteinase K, 37 ℃ process 30-60min, then in final concentration 1% ( mass percent concentration) in SDS solution at 65°C for 1-2 hours, centrifuge (at room temperature) to take the supernatant, and extract once with an equal volume of chloroform to remove the protein. If the interface between water and chloroform solution is not clear after extraction, Consider re-extracting ...
Embodiment 2
[0059] Specificity analysis of embodiment 2 RealAmp detection
[0060] Other rice pathogens used for specific analysis are rice blast (Magnaporthe grisea), sheath blight (Thanatephorus cucumeris), rice bakanae (Gibberella moniliformis), bacterial blight (Xanthomonas oryzae pv.oryzae), bacterial Stripe (Xanthomonas oryzae pv.oryzicola), and the insecticidal fungus Metarhizium anisopliae (see Table 1 for details).
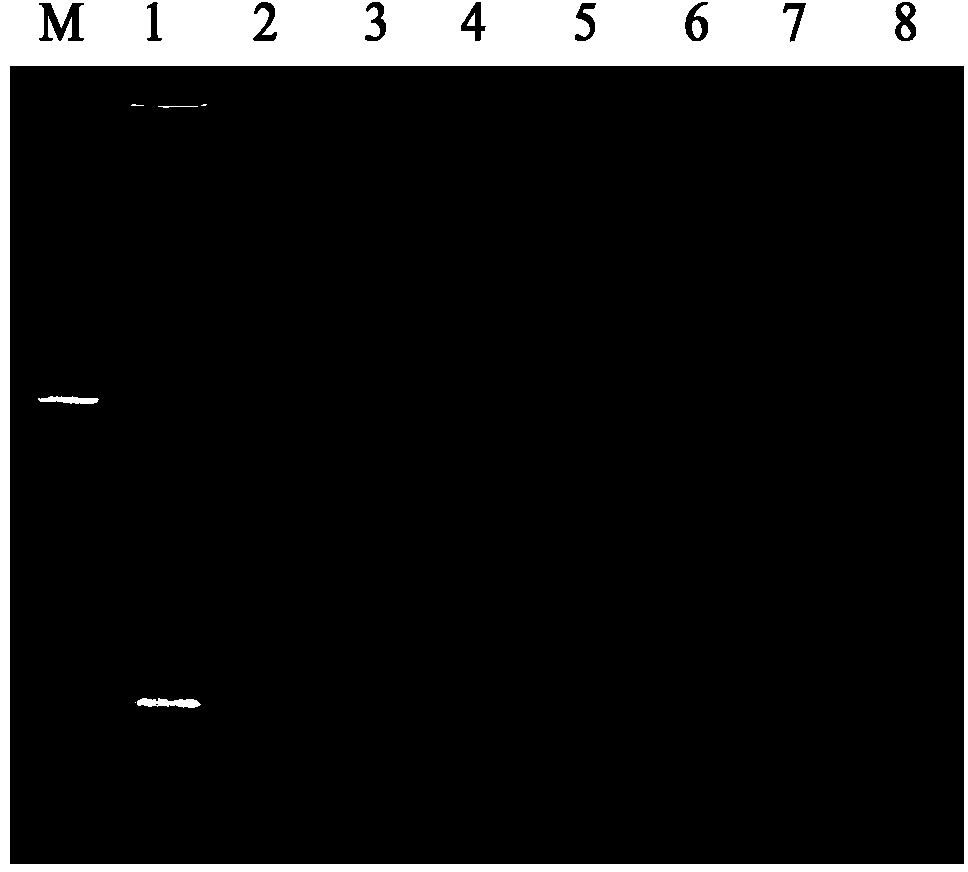
[0061] The product electrophoresis detection results after constant temperature amplification showed that only the template was rice smut (Ustilaginoidea virens) genomic DNA showing gradient bands, and the rest of the pathogenic DNA had no gradient bands (see figure 1 ); All bacterial strains adopt the nested PCR detection of rice false smut bacterium, and the result shows, only has the specific band of 232bp amplified by the rice false smut genomic DNA, and other controls have no amplified product (see figure 2 ); the ESE-Quant Tube Scanner instrument has only one...
Embodiment 3
[0066] The analysis of embodiment 3 seed carrier detection sensitivity and standard curve construction thereof
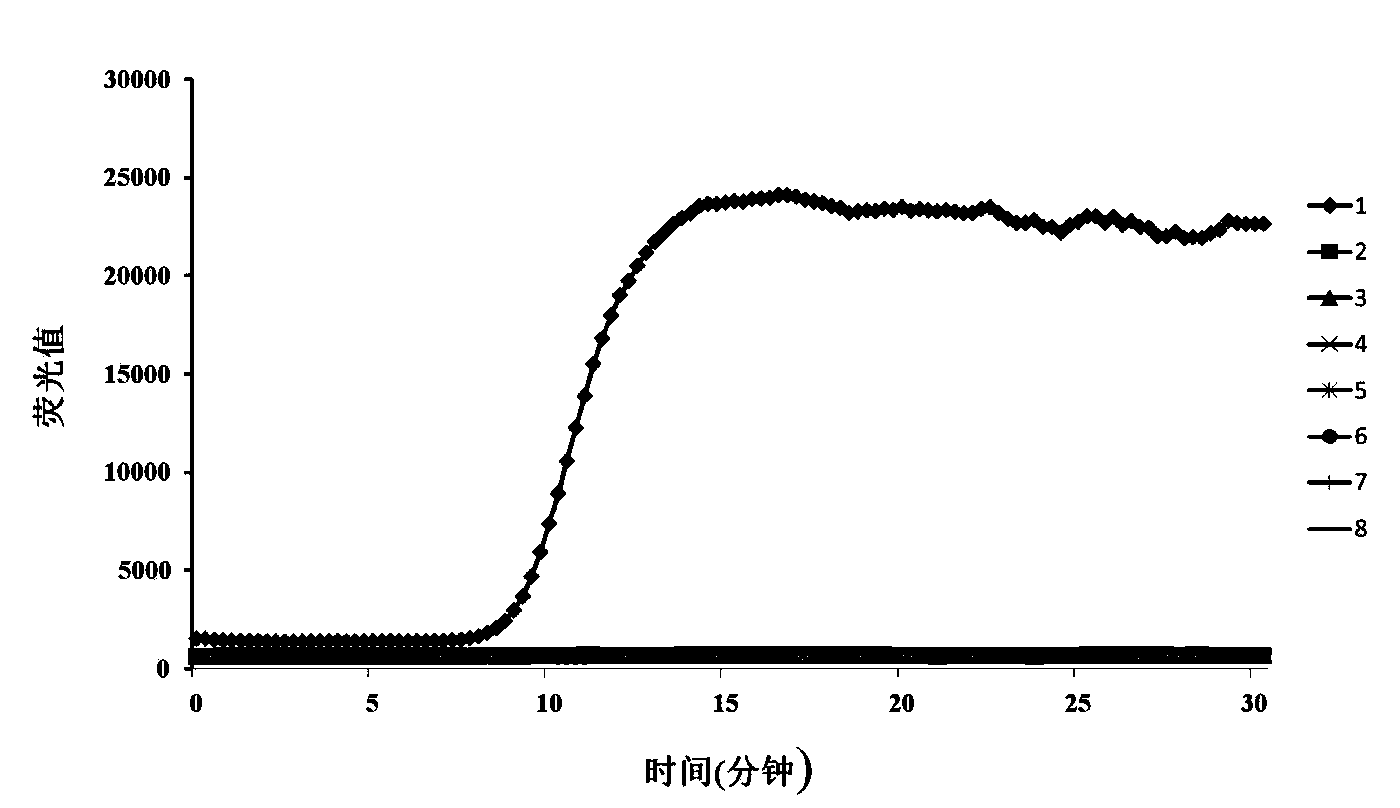
[0067] Use the pMD-18-T-UV plasmid DNA containing the RealAmp amplification region as a template, and use the healthy testa DNA as water to dilute the pMD-18-T-UV plasmid DNA sequentially from 38ng / μl by 10-fold gradient serial dilution method to 3.8×10 -6 ng / μl, sterile water was used as negative control. The results showed that, RealAmp can be obtained from pMD-18-T-UV plasmid DNA concentration is 38ng / μl~3.8×10 -4 The products amplified from template DNA with 6 orders of magnitude such as ng / μl were all green after staining with SYBR GreenI, and the negative control was colored orange, indicating that the lower limit of detection of Aspergillus oryzae DNA by RealAmp is about 3.8×10 -4 ng / μl (see Figure 4 );
[0068] The amplification curves of ESE-Quant Tube Scanner showed a highly linear relationship between different concentrations of starting template DNA...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com