Method for rapidly detecting and identifying Listeria bacteria
A technology for Listeria monocytogenes and Listeria monocytogenes is applied in the field of molecular biological detection, which can solve the problems of high cost, high work intensity, complicated steps and time, and achieve the effects of improving efficiency, saving time and having good application prospects.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
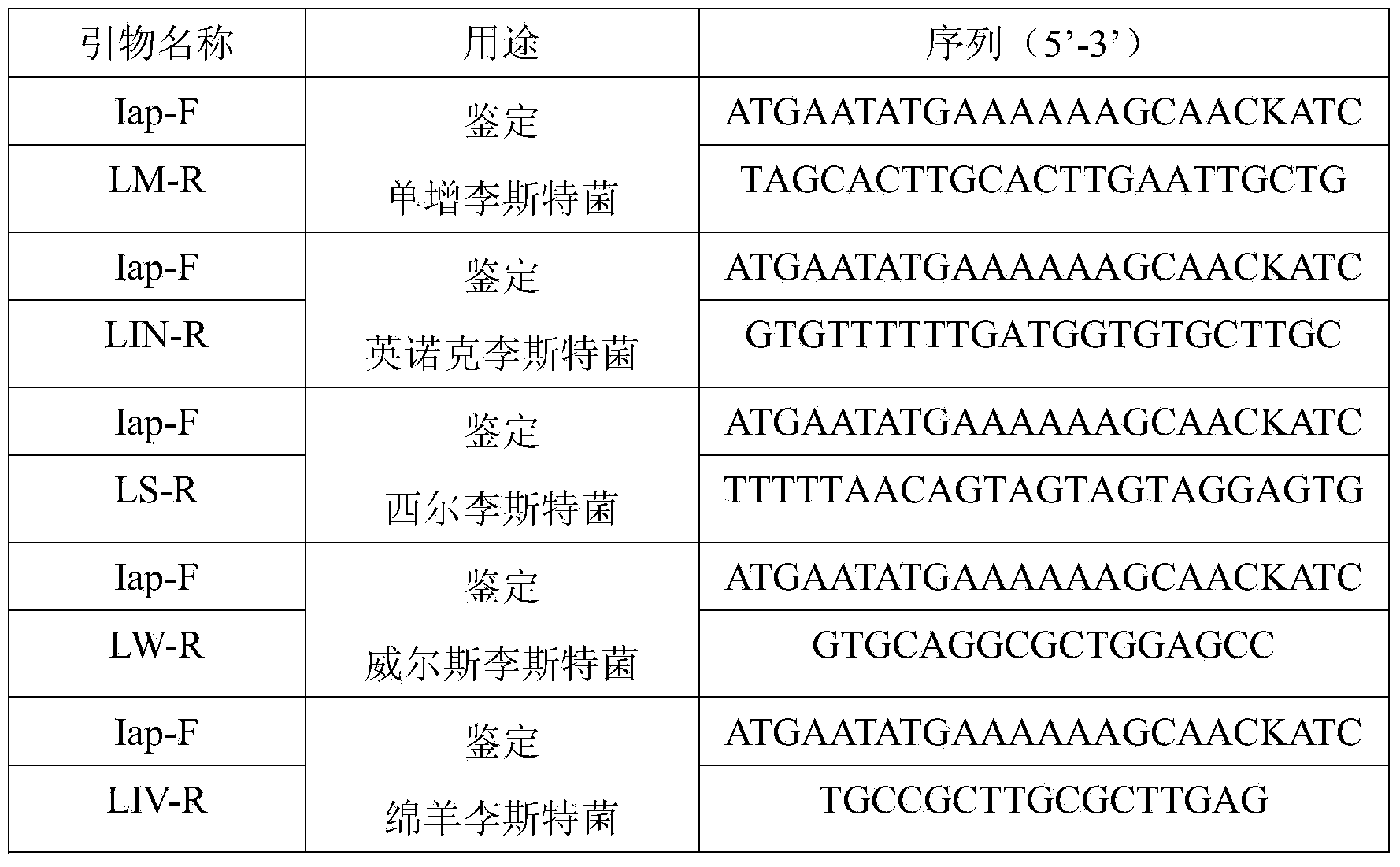
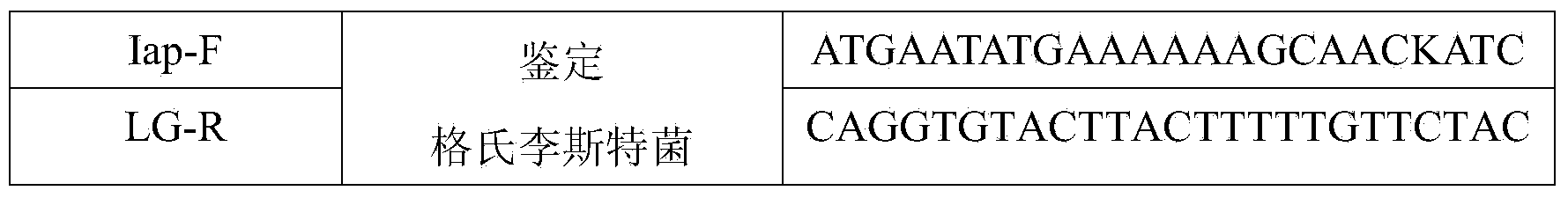
[0046] Example 1 PCR detection method for specificity evaluation of different strains
[0047] 1 Materials and methods
[0048] 1.1 Materials
[0049] 1.1.1 Medium
[0050] Listeria Selective Enrichment Solution (LB1, LB2), Tryptone Soy Broth (TSB), Tryptone Soy Agar (TSA) were purchased from Beijing Land Bridge Technology Co., Ltd.; Listeria Selective Agar (PALCAM), Brain heart infusion (BHI), antibiotics, etc. were purchased from OXOID, UK;
[0051] 1.1.2 Main reagents
[0052] PCR Premix (including rTaq, 10×buffer(+Mg 2+ ), and dNTP Mixture), DNA Marker (DL2000bp), and DNA Recovery Kit were purchased from Dalian Bao Biological Engineering Co., Ltd.; Biospin Bacterial Genomic DNA Extraction Kit was purchased from Hangzhou Bioer Company; agarose, EB (ethidium bromide) Bought from Tiangen Biotechnology Company; biochemical identification kits were purchased from Beijing Land Bridge Co., Ltd.; other reagents: anhydrous ethanol, 95% ethanol, glycerin, sodium chloride, etc. ...
Embodiment 2
[0101] Example 2 The experiment of detecting Listeria in the environment represented by fresh pork samples
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com