An engineering bacterium for knocking out pyruvate formate lyase gene and its application
A pyruvate formic acid and lyase technology, applied in the biological field, can solve the problems of substrate glycerol waste, inhibition of cell growth, and reduced production of 1,3-propanediol, and achieve the effects of reduced formic acid production, reduced toxic effects, and improved levels
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0022] Example 1: Construction of a Klebsiella pneumoniae mutant strain in which the key gene of formic acid metabolism pathway-pyruvate formate lyase flpB gene is inactivated.
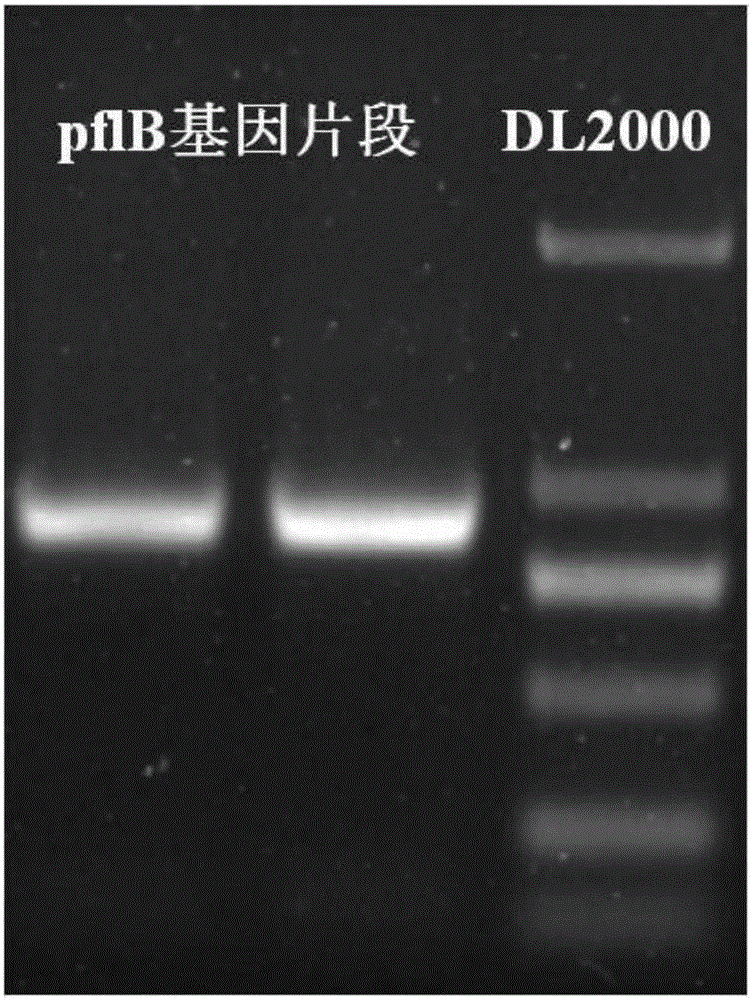
[0023] (l) Clone the partial sequence of pyruvate formate lyase gene pflB
[0024] Design primers for PCR amplification of part of the pyruvate formate lyase flpB gene sequence. The primer sequences are as follows: upstream primer pflB-F: taggtacctgaaagacaaattcgcccag and downstream primer pflB-R: gagagctccatgcgatccattacttcgt. Using wild-type Klebsiella pneumoniae (deposited in the Chinese Type Culture Collection, deposit number: CCTCCM2011075) genomic DNA as a template, under the guidance of primers pflB-F and pflB-R, PCR amplification pyruvate formic acid cleavage For the partial sequence of the enzyme pflB, the PCR amplification conditions are: first 95℃ for 3 min; then 94℃ for 1 min, 50℃ for 1 min, 72℃ for 1 min, a total of 32 cycles; finally 72℃ for 10 min. After the reaction, the PCR amplified produc...
Embodiment 2
[0029] Example 2: Detection of the activity of Klebsiella pneumoniae mutant strains in which the pyruvate formate lyase flpB gene was inserted and inactivated.
[0030] The Klebsiella pneumoniae mutant strain in which the pyruvate formate lyase flpB gene of Example 1 was inserted and inactivated was tested for the activity of pyruvate formate lyase. The wild-type Klebsiella pneumoniae was used as a control, and the specific methods included The following steps:
[0031] (l) Inoculate the Klebsiella pneumoniae mutant strain with inactivated pyruvate formate lyase flpB gene in 100 mL of medium (each liter of water contains 20 g of glycerol, 10 g of tryptone, 5 g of yeast powder, 5 g of NaCl, pH 7.0, Sterilize at 120°C for 20 minutes), culture with shaking at 37°C for 6-12 hours, and collect samples by centrifugation every 2 hours;
[0032] (2) Suspend and wash the bacteria twice with 100 mL phosphate buffer (0.1M, pH7.5);
[0033] (3) Use 2.5mL phosphate buffer (0.1M, pH7.5) to suspend...
Embodiment 3
[0037] Example 3: Fermentation of 1,3-propanediol by Klebsiella pneumoniae mutant strain knocked out of pyruvate formate lyase flpB gene
[0038] (1) Medium
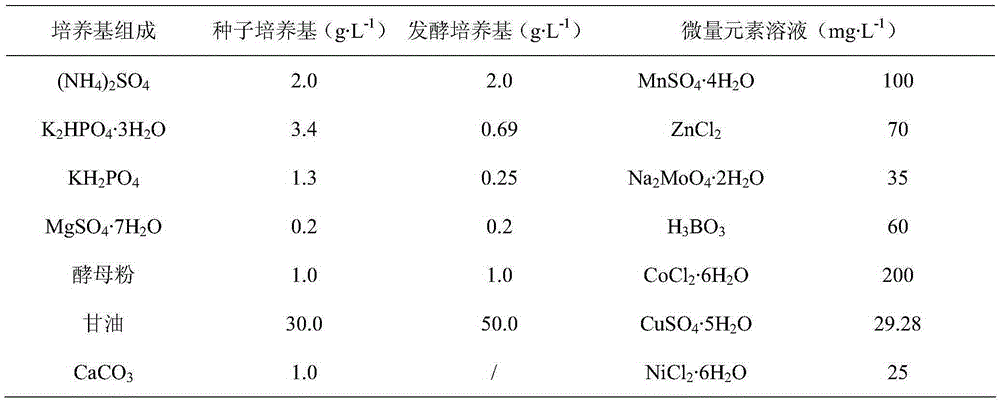
[0039] LB medium (g·L -1 ): Yeast powder 5, peptone 10, NaCl10, agar 10, adjusted to pH 7.0, used for short-term preservation and activation of Klebsiella strains. See Table 1 for the composition of seeds and fermentation medium:
[0040] Table 1: Medium composition
[0041]
[0042]
[0043] (2) Training method
[0044] (i) Seed activation: Klebsiella pneumoniae mutant strains and wild bacteria that knocked out the pyruvate formate lyase flpB gene in Example 1 stored in glycerol tubes were respectively inoculated to the LB medium slope for activation, and the temperature was 37°C Cultivate for 12 hours to activate the seeds.
[0045] (ii) Seed culture: a 250mL Erlenmeyer flask is sealed with 9 layers of gauze, 100mL seed culture medium filled with liquid, connected to the slant moss (activated seed in step i), and aerobic seed c...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com