Method for detecting CHO cell proliferation by using optical analysis and application thereof
A cell proliferation and optical analysis technology, applied in the field of non-invasive optical analysis to detect Chinese hamster ovary cell proliferation, can solve the problems of compound toxicity, reduced accuracy, staining dispersion, etc., and achieve good reproducibility, good repeatability, and operation. convenient effects
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
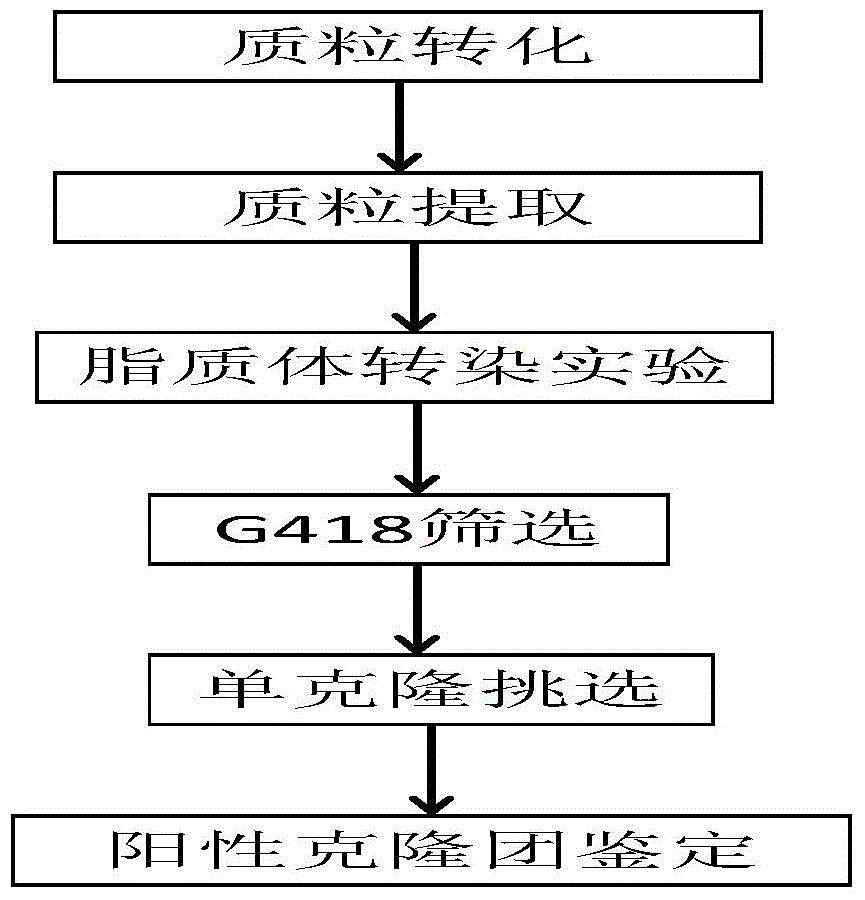
Embodiment 1
[0060] Preparation of pEGFP-N2
[0061] (1) Take a competent cell of Escherichia coli DH5α, put it on ice to thaw for 30 minutes, then take 1 μl of pEGFP-N2 plasmid and add it to the competent cell, and mix well by finger flicking. After 30 min of ice bathing, 42°C water bath for 90 sec, repeat the ice bath for 2 min for transformation. After the transformation is completed, take 0.8ml of SOC medium to suspend the bacteria, and resume culture at 37°C for 1 hour on a shaker at 180rpm.
[0062] (2) Take the recovered bacterial solution, centrifuge and resuspend the pellet, take 10ul and spread it on the resistant LB plate, and culture overnight at 37°C. Pick clones with good growth potential, inoculate them in LB liquid medium containing resistance, and cultivate overnight at 37°C on a shaker at 180rpm.
[0063] (3) Extract the plasmid for sequencing. After the sequencing result is correct, shake the bacteria, use the OMGA plasmid extraction kit to extract the endotoxin-free p...
Embodiment 2
[0069] Liposome lipofectimene3000 transfection and its screening
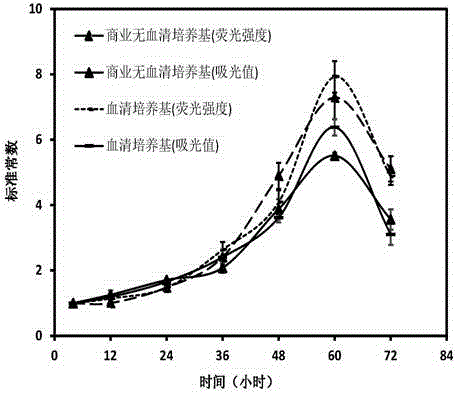
[0070] (1) The revived CHO cells were inoculated into T-flask culture flasks containing 6ml of growth medium (10% FBS+1% Penicillin-Streptomycin Solution+89% DMEM / F12, hereinafter referred to as 10% FBS medium group), at 37 °C, 5% CO 2 Cultured in a constant temperature cell incubator until the cell density reached 90%. After several generations of culture, inoculate 1×10 5 cells in a 24-well plate at 37°C, 5% CO 2 Culture at constant temperature until the cell growth density reaches 70%.
[0071] (2) Refer to the procedure of lipofectimene3000 kit for transfection operation. Mix lipofectimene3000 reagent and opti-MEM medium uniformly to obtain solution A; mix 1ug pEGFP-N2, opti-MEM medium and P3000 reagent uniformly to obtain solution B. Equal volumes of solutions A and B were mixed in a 1.5ml centrifuge tube, incubated for 5 minutes and then added to a 24-well plate for culture. After 48 hours of transf...
Embodiment 3
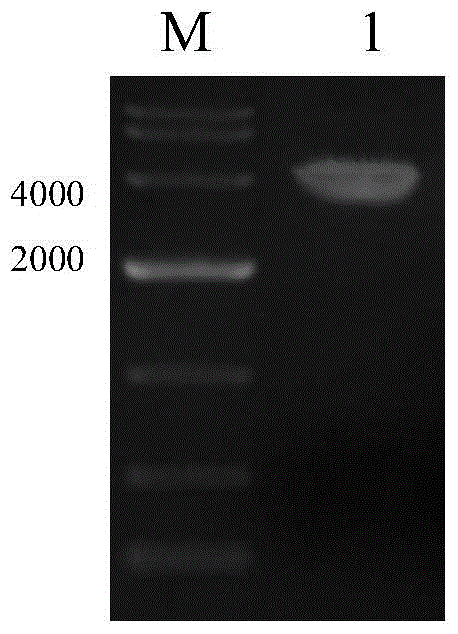
[0075] PCR identification of resistant cell clones
[0076] In order to identify whether the target gene GFP is fully integrated into the cell genome, the whole cell genome was extracted using the Takara Genome Extraction Kit.
[0077] (1) Using the genome obtained from lysed cells as a template. The genome template of untransfected CHO-K1 cells was used as a negative control.
[0078] The primer sequences are as follows:
[0079] pEGFP-N2-F2: 5'-CTGGTCGAGCTGGACGGCGACG-3'
[0080] pEGFP-N2-R2: 5'-CACGAACTCCAGCAGGACCATG-3'
[0081] The reaction system is shown in Table 2:
[0082] Table 2 PCR reaction system for cell identification
[0083]
[0084] All experimental operations should be carried out under sterile conditions to prevent cross-contamination.
[0085] (2) Amplify the target fragment according to the PCR program set below.
[0086] PCR program settings:
[0087]
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com