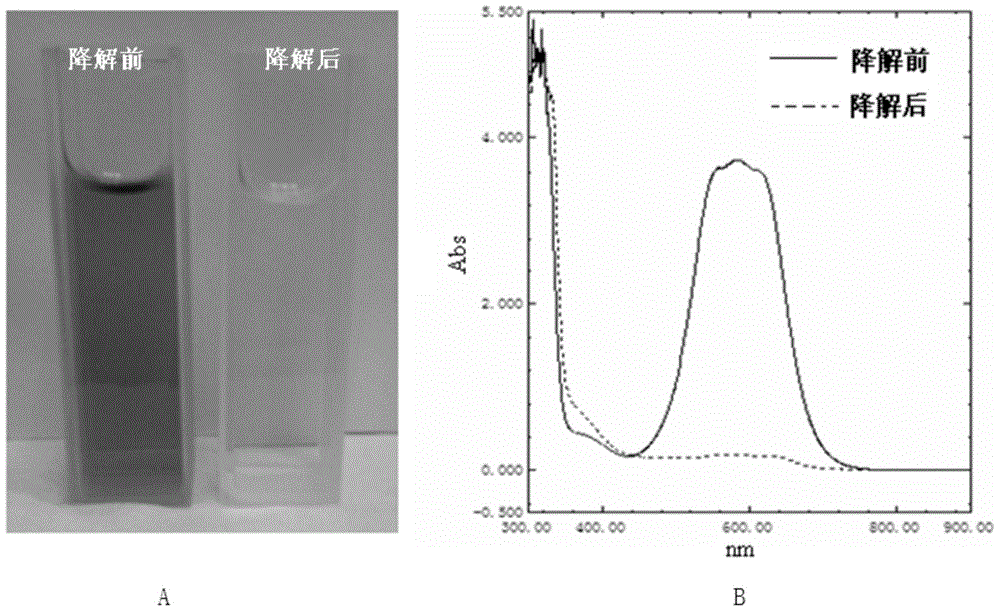
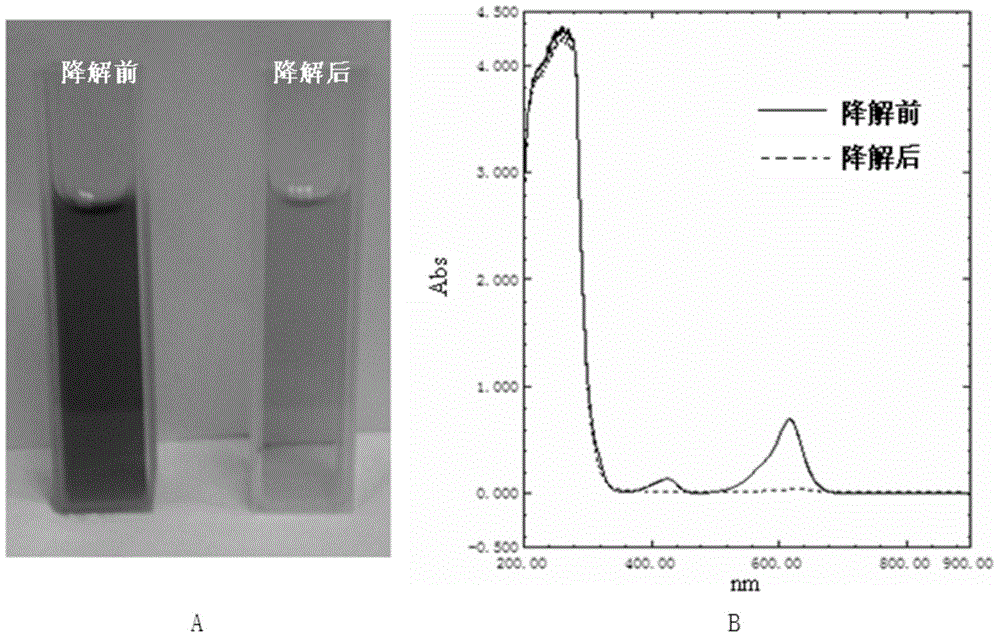
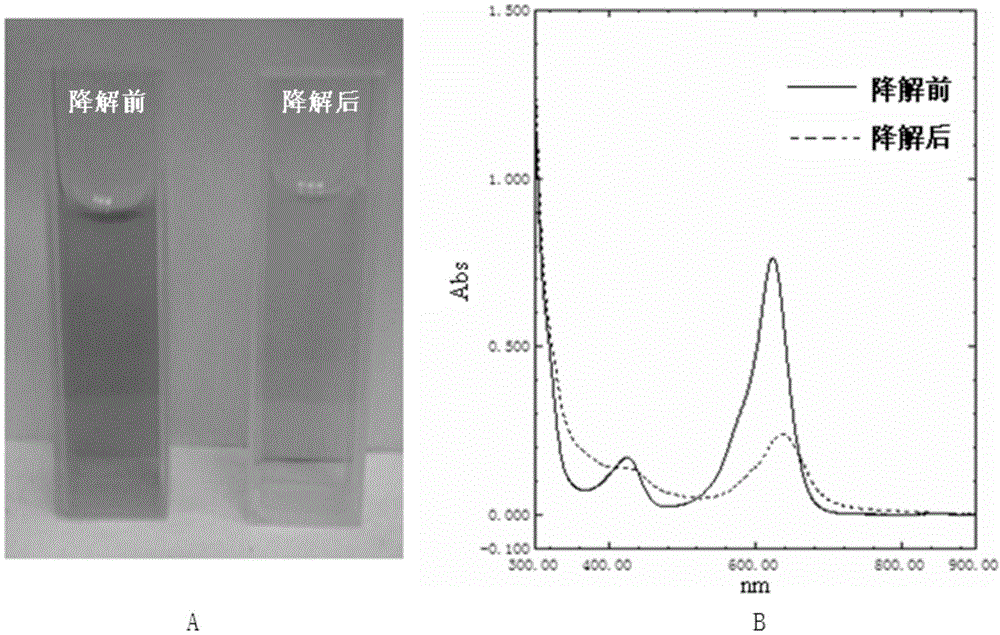
Method for degrading triphenylmethane dye by utilizing recombinational lipoxygenase
A technology of lipoxygenase and triphenylmethane, applied in the biological field, can solve the problems of unsatisfactory treatment effect and difficult degradation of biological treatment system
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0030] Example 1: Cloning of Pseudomonas aeruginosa ATCC27853 lipoxygenase gene (LOX-27853)
[0031] The bacterial cells of Pseudomonas aeruginosa ATCC27853 were collected by centrifugation, and the genomic DNA of Pseudomonas aeruginosa ATCC27853 was extracted with the Shanghai Sangon Genomic DNA Extraction Kit.
[0032] According to the registered Pseudomonas lipoxygenase gene (No.CP006832.1) in the Genebank database, two primers were designed:
[0033] Upstream primer F-1: 5'-ATGAAACGCAGGAGTGTGCTCTTG-3' (SEQ ID NO.3);
[0034] Downstream primer R-1: 5'-TCAGATATTGGTGCTCGCCGGGATC-3' (SEQ ID NO.4);
[0035] In a 50 μl system, the final concentration of each primer is 1 μM, the final concentration of dNTPs is 0.2 mM, 10 ng of genomic DNA of Pseudomonas aeruginosa ATCC27853 strain, 2 U of Pfu DNA polymerase. The amplification program was 94°C for 3min; 30×(94°C for 40s, 53°C for 50s, 72°C for 90s); 72°C for 10min. Agarose gel electrophoresis, gel cutting, recovery by Shanghai ...
Embodiment 2
[0036] Embodiment 2: the construction of Pseudomonas aeruginosa ATCC27853 lipoxygenase gene (LOX-27853) prokaryotic expression vector (attached figure 1 )
[0037] According to the obtained laccase gene sequence, two primers were designed, the upstream primer plus the SacI recognition sequence, and the downstream primer plus the XhoI recognition sequence (the underlined part is the restriction enzyme recognition sequence):
[0038] Upstream primer F-2: 5′-CGC GAGCTC ATGAAACGCAGGAGTGTGCTCTTG-3' (SEQ ID NO. 5)
[0039] Downstream primer R-2: 5′-CCG CTCGAG TCAGATATTGGTGCTCGCCGGGATC-3' (SEQ ID NO. 6)
[0040] Add each component according to the following PCR system to amplify the LOX gene:
[0041]
[0042] The PCR program is: 94°C for 3min; 30×(94°C for 40s; 53°C for 50s; 72°C for 90s); 72°C for 10min.
[0043] Purify the PCR product with Shanghai Sangon PCR Product Purification Kit, add SacI, XhoI double enzyme digestion, inactivation, ethanol precipitation, ddH 2 O was...
Embodiment 3
[0044] Embodiment 3: Fermentative production of Pseudomonas aeruginosa ATCC27853 lipoxygenase gene (LOX-27853) in Escherichia coli
[0045] The fermentation production of Pseudomonas aeruginosa ATCC27853 lipoxygenase gene (LOX-27853) in Escherichia coli involves the following three media:
[0046] (1) Solid plate medium: 10g / L tryptone, 5g / L yeast extract, 10g / L sodium chloride, 15g / L agar powder
[0047] (2) Seed liquid medium: 10g / L tryptone, 5g / L yeast extract, 10g / L sodium chloride
[0048] (3) Fermentation medium: 10g / L lactose, 0.5g / L glucose, 5g / La glycerol, 3.4g / L KH 2 PO 4 , 1.2g / L MgSO 4 , 10g / L Enzymatic Sodium Caseinate, 5g / L Yeast Extract, 8.95g / L Na 2 HPO 4 .12H 2 O, 1.42g / L Na 2 SO 4 ,2.67g / L NH 4 Cl and 1ml trace element mixture.
[0049] (4) Trace element mixture: 50μM FeCl 3 .6H 2 O, 20 μM CaCl 2 .2H 2 O, 10 μM MnCl 2 .4H 2 O, 10 μM ZnSO 4 .7H 2 O, 2 μM CoCl 2 .6H2O, 2μM CuCl 2 ,2μM NiCl 2 ,2μM Na 2 SeO 3 ,2μM H 3 B 4 o 7 .
[0050]...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com