Gene chip and method for detecting pathogenic microorganisms of porcine epidemic disease
A pathogenic microorganism and gene chip technology, applied in the field of swine blight pathogenic microorganism detection, can solve the problems of time-consuming, labor-intensive, poor sensitivity and specificity, and achieve the effect of improving detection efficiency, strong specificity and saving time.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0057] Embodiment 1: the preparation of gene chip
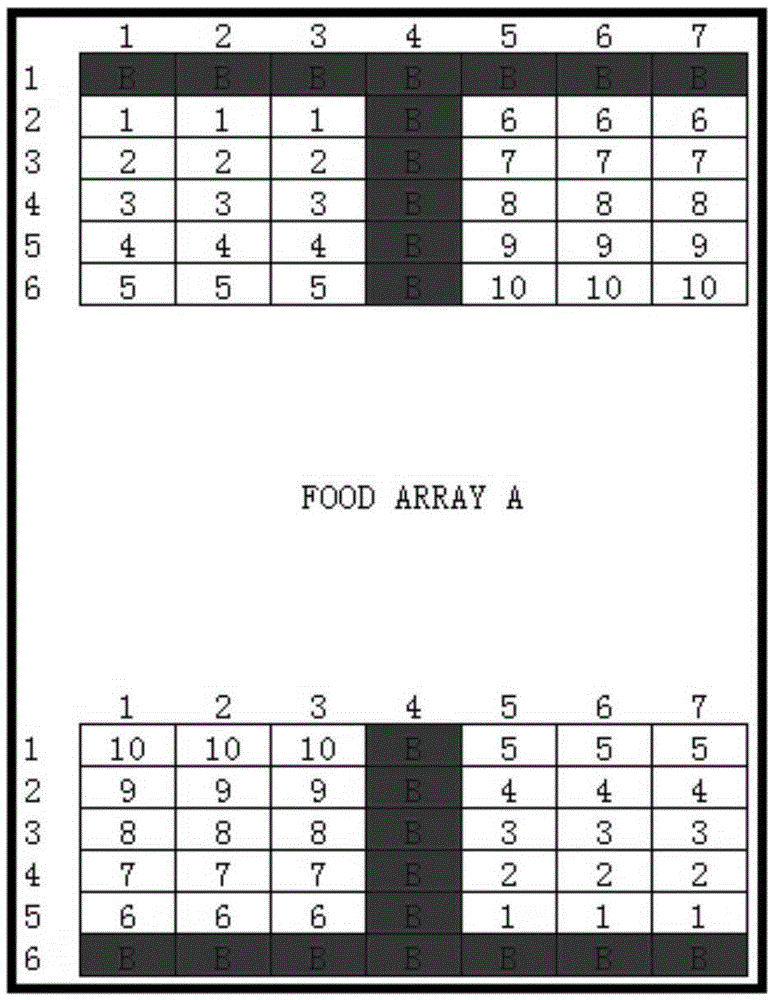
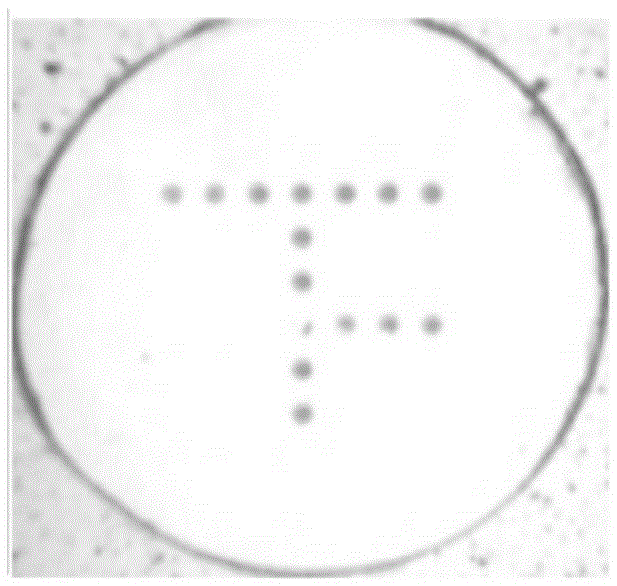
[0058] Spot the designed oligonucleotide probes corresponding to the pathogenic microorganisms to be detected on positively charged glass slides (AMI Company, U.S.), and the spotted glass slides are exchanged under a long-wave ultraviolet lamp. Integrate for about 10 minutes, and at the same time, 0.2 μL of positive quality control labeled with biotin is also spotted on each glass slide. The distribution of positive quality controls is in the shape of a "T", and the oligonucleotide probes corresponding to the pathogenic microorganisms to be tested are distributed in a microarray. The pattern diagram of the gene chip is as follows: figure 1 shown.
[0059] The sequence of the oligonucleotide probe corresponding to the pathogenic microorganism to be detected is as follows:
[0060] (1) Probe for Actinobacillus pleuropneumoniae
[0061] SEQ ID NO.1: 5'gcaAAGGTAATGATATTCTAAgaggt
[0062] (2) Probe for classical swine fever v...
Embodiment 2
[0080] Example 2: Screening of primers for specific amplification of specific conserved nucleotide sequences of pathogenic microorganisms to be detected
[0081] Effective design of primers for specific amplification is the most critical link in determining the success of the experiment. Although there are some software for designing primers and some basic principles for designing primers, according to these design software and design principles, multiple different primer sequences can be designed, and the use of different primer sequences will produce different effects. The replacement or increase or decrease will have an important impact on the test results. Therefore, the design of amplification primers cannot be obtained by conventional software design. In this example, it is used to specifically amplify Actinobacillus pleuropneumoniae, classical swine fever virus, Haemophilus parasuis, swine influenza virus, Mycoplasma hyopneumoniae, porcine circovirus type Ⅱ, porcine pa...
Embodiment 3
[0163] Example 3: Specific verification of the gene chip
[0164] Sample: Specific virus and bacterial culture fluid (specific viruses and bacteria are respectively: swine fever virus, porcine blue ear virus, Actinobacillus pleuropneumoniae, Haemophilus parasuis and porcine pseudorabies virus).
[0165] 1 sample processing
[0166] Centrifuge 100mL of bacterial culture solution, resuspend in normal saline, add lyoprotectant (5% skim milk powder), freeze-dry and store; take 10mL of virus culture solution, add lyoprotectant (5% skim milk powder), freeze-dry Store dry.
[0167] 2 total RNA extraction
[0168] Freeze-dried samples of bacteria and virus were taken, resuspended in PBS to 50-100 μL, and a total RNA extraction kit (TIANGEN, Germany) was used to operate according to the instructions.
[0169] 3RT-PCR amplification
[0170] The primers optimized in Example 2 were used as specific amplification primers, and the specific amplification primers were biotin-labeled (labe...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com