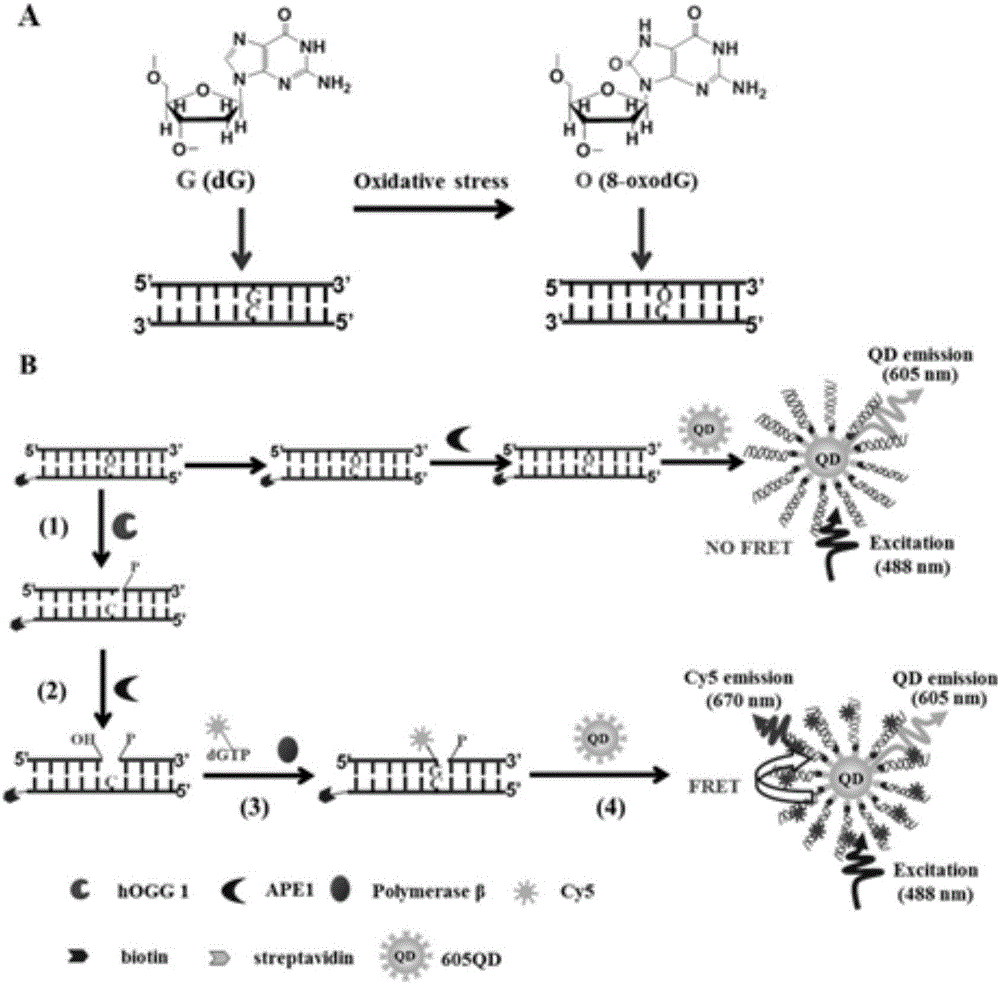
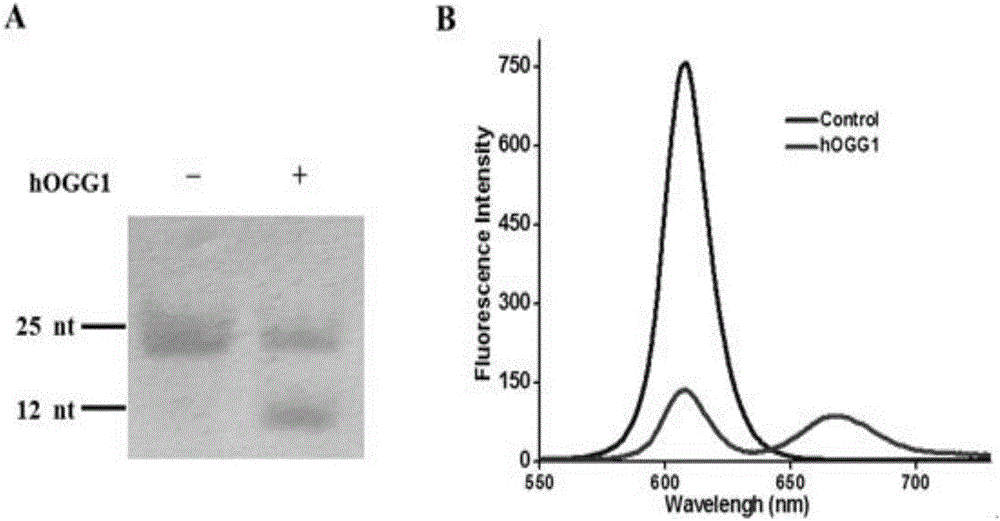
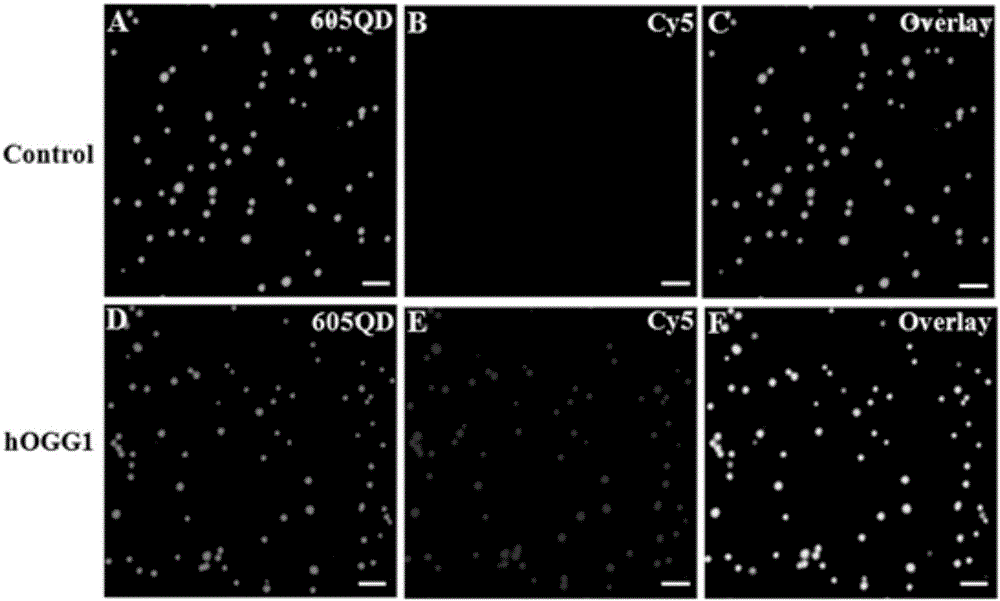
Method for detecting DNA (deoxyribonucleic acid) glycosylase activity on basis of single quantum dot level
A glycosylase, level detection technology, applied in the field of biological analysis, can solve the problems of complex operation, low detection limit, long time consumption, etc., to achieve the effect of good specificity, high specificity and reducing non-specificity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0035] Preparation of the incubation buffer: 100 millimoles per liter of tris(hydroxymethyl)aminomethane-hydrochloric acid (Tris-HCl), 10 millimoles per liter of ammonium acid, 3 millimoles per liter of magnesium chloride, 0.83 nanosulfide per liter Liter of quantum dots (605QDs), pH 8.0.
[0036] Preparation of anti-quench buffer: 67 millimoles per liter of glycine-potassium hydroxide (pH 9.4), 2.5 millimoles per liter of magnesium chloride, 50 micrograms per milliliter of bovine serum albumin, 1 mg per milliliter of glucose oxidation Enzyme, 0.04% mg / ml catalase, 0.4% (mass / volume) D-glucose.
[0037] Cell extract preparation: Human lung adenocarcinoma cell (A549) medium is Dulbecco's modified Eagle's medium (DMEM) containing 10% fetal bovine serum (FBS) and 1% penicillin-streptomycin. Cultivate in an incubator with 5% carbon dioxide and 37 degrees. When the cells grow to the logarithmic growth phase, they are digested with trypsin, washed twice with phosphate buffered saline (...
Embodiment 2
[0043] 2.1 Experimental verification of principle
[0044] In order to verify the feasibility of DNA glycosylase hOGG1 in extracellular base excision repair, the inventors tested and analyzed the repair reaction products, and the results are as follows figure 2 Shown. First, the present invention uses non-denaturing polyacrylamide gel (PAGE) electrophoresis for verification analysis. From figure 2 It can be seen from A that when there is no DNA glycosylase hOGG1, there is only a 25bp band, indicating that the excision repair reaction did not occur. When there is DNA glycosylase hOGG1, two bands can be seen, 25bp and 12nt in length, indicating that one of the DNA strands is cut and a small 12nt fragment is produced. The above results show that DNA glycosylase hOGG1 can specifically recognize and excise 8-oxoguanine (8-oxoG), and under the cleavage of apurinic endonuclease-1 (APE1), the abasic site (APsite ) Leaves a gap in nucleotides. Cy5-dGTP is added and DNA polymerase β p...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com