CBLB502-Fc fusion protein and preparation method thereof
A technology of CBLB502 and fusion protein, which is applied in the fields of peptide/protein components, chemical instruments and methods, hybrid peptides, etc., can solve the problems of complex purification process, difficult removal of heat source or endotoxin, and achieve simplified protein purification process and improved stability Sexuality, the effect of great application prospects
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
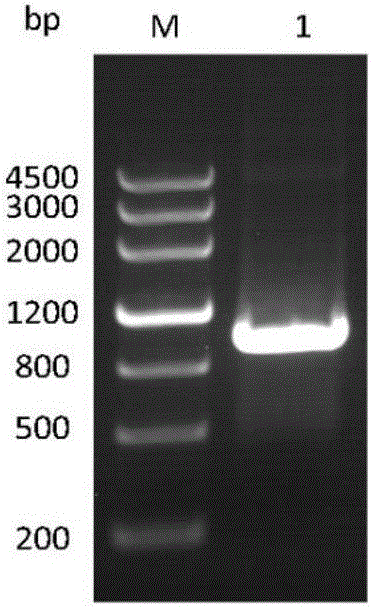
[0053] The amplification of embodiment 1CBLB502 gene
[0054] Primers 502F and 502R were designed according to the CBLB502 gene sequence, and the target gene fragment was amplified by DNA polymerase PCR using the vector pUC57-CBLB502 as a template. Among them, the base sequence (5'-3') of 502F is GATATCCCTAGGCCACCATGGAA, and the restriction sites are AvrII and EcoRV; the base sequence (5'-3') of 502R is GGATCCCAGCAGGCTCAGGACG, and the restriction site is BamHI.
[0055] PCR reaction system (50 μL) includes: 40ng template DNA sample, 1.5 μL forward primer set 502F (0.3 μM), 1.5 μL reverse primer set 502R (0.3 μM), 25 μL 2×PCR buffer, 10 μL dNTP (2 mM), 1 μL KODFX DNA polymer Enzyme (1U), ddH 2 O supplemented to 50 μL.
[0056] The PCR reaction conditions included: 94°C pre-denaturation for 2 min, 98°C denaturation for 10 s, 65°C annealing for 30 s, 68°C extension for 1 min, 30 cycles, and then 68°C extension for 7 min. The PCR amplification product was subjected to 1.0% agar...
Embodiment 2
[0057] Example 2 Construction and identification of pUC57-CBLB502-Fc plasmid
[0058] The target gene fragment and pUC57-Fc were double-digested with restriction endonucleases AvrII and BamHI, and the digested products were detected by 1.0% agarose gel electrophoresis, and the target fragment was recovered with a DNA gel recovery kit after gel cutting. Transform competent Escherichia coli after ligation with T4 DNA ligase, select positive single clones, verify and sequence with primers P57YF and P57YR. The base sequence (5'-3') of P57YF is CGCCAGGGTTTTTCCCAGTCAC, and the base sequence (5'-3') of P57YR is GCAGGACAGGGAGGACAAGTG.
[0059] The positive colony identification method is as follows: select positive monoclonal to ampicillin-resistant LB medium, shake the bacteria on a constant temperature shaker at 37°C for 6 hours, and take the bacterial liquid for PCR verification. PCR reaction system (20 μL): 1 μL bacterial liquid, 1 μL P57YF, 1 μL P57YR, 10 μL 2×TaqMasterMix, 7 μL...
Embodiment 3
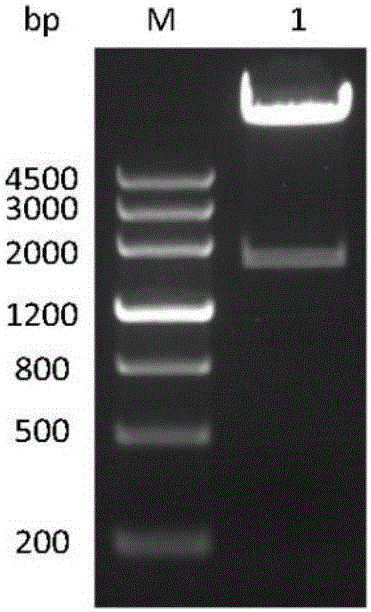
[0062] Example 3 Construction and identification of pcDNA3.1-CBLB502-Fc plasmid
[0063] pcDNA3.1 and pUC57-CBLB502-Fc were digested with restriction endonucleases EcoRV and HindIII, and the CBLB502-Fc gene fragment obtained after digestion was cloned into a vector, transformed into Escherichia coli competent cells, and positive single clones were selected and used Restriction digestion was verified and sent for sequencing, and the constructed plasmid was named pcDNA3.1-CBLB502-Fc. CBLB502-Fc-specific bands can be seen at about 1800bp, such as figure 2 shown, consistent with the expected results. The obtained recombinant plasmid was sequenced, and the sequencing result was correct, and the pcDNA3.1-CBLB502-Fc plasmid was constructed successfully.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com