RT-PCR detection method, primer and probe as well as kit of Zika virus, dengue virus and chikungunya virus
A technology of dengue virus and chikungunya fever, which is applied in the field of molecular biology detection of insect-borne infectious diseases, can solve the problems of detection products that have not been reported, and achieve the goals of shortening detection time, improving detection efficiency and saving costs Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0059] Primer and probe design
[0060] The design of primers and probes mainly refers to the gene sequences of all Zika virus, dengue virus and chikungunya virus in the GenBank database, using the BLAST tool on the NCBI website, DNAStar, ABI’s PrimerExpress and other software and self-designed analysis The software analyzes the conserved sequences of Zika virus, dengue virus and chikungunya virus, and designs specific primers and probes based on the obtained conserved sequences, analyzes the interaction between primers, optimizes and screens primer-probe combinations, Minimize the cross-reaction between flaviviruses to ensure the high specificity, high sensitivity and high genotype coverage of the reagent. The resulting primers and probes are shown in Table 1:
[0061] surface Zika virus, dengue virus and chikungunya virus and internal standard detection primer probe sequences
[0062]
[0063] The 5' end of SEQ ID No.3 is marked with a fluorescent reporter group, and...
Embodiment 2
[0065] Detection of Zika virus, dengue virus, chikungunya virus triple real-time fluorescent RT-PCR in clinical samples
[0066] 1. Sample processing: Take a certain amount of serum sample and add 3 times the volume of methanol, mix well and let it stand for five minutes, centrifuge at 3000r / min for 15min, and select the supernatant for nucleic acid extraction of viral RNA. Viral RNA was extracted using the ReansyMini Kit from QIAGEN, Germany, and extracted according to the kit instructions to obtain viral RNA. 10-fold serial dilutions were performed with LB buffer and labeled.
[0067] 2. Reagent preparation:
[0068]
[0069] RT-PCR reaction solution consists of: RT-PCR buffer, magnesium chloride, deoxyribonucleotide triphosphate mixture and PCR enhancer; PCR buffer includes Tris-HCl (pH8.0), ammonium sulfate, potassium chloride and Triton X-100; the PCR enhancer is selected from one or more of tetramethylammonium chloride, carnitine, trehalose and non-ionic detergent N...
Embodiment 3
[0089] Comparison of Analytical Sensitivity of Zika Virus, Dengue Virus, Chikungunya Virus Triple Real-Time Fluorescent RT-PCR and Single-plex Conventional Reagents
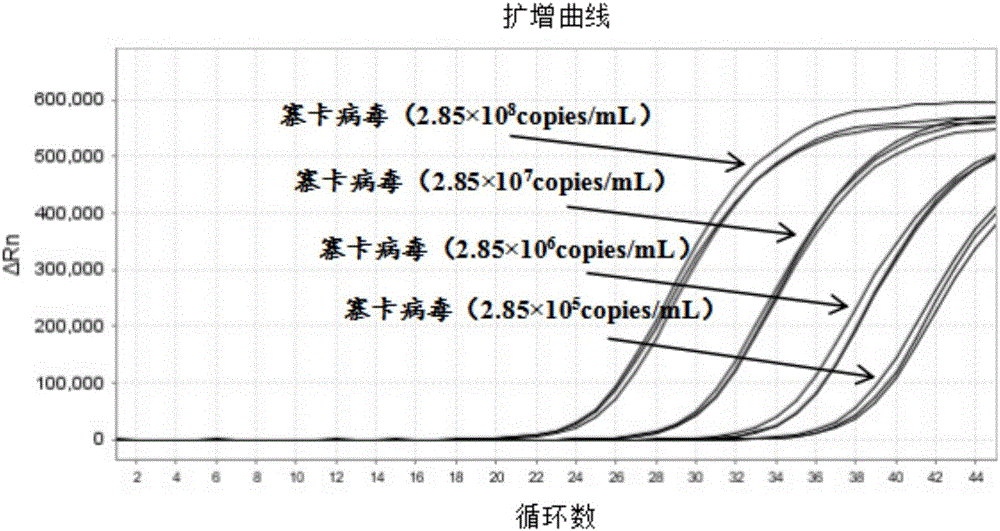
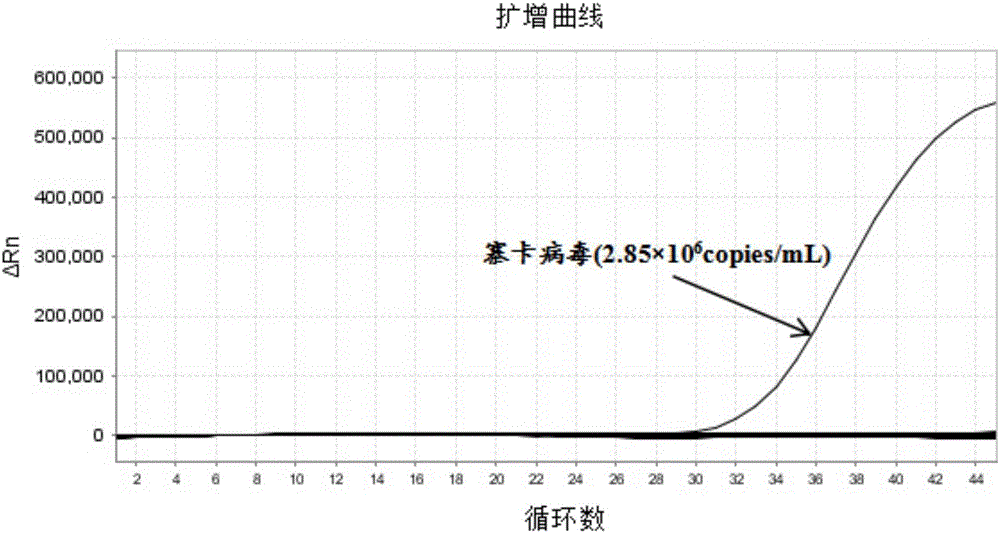
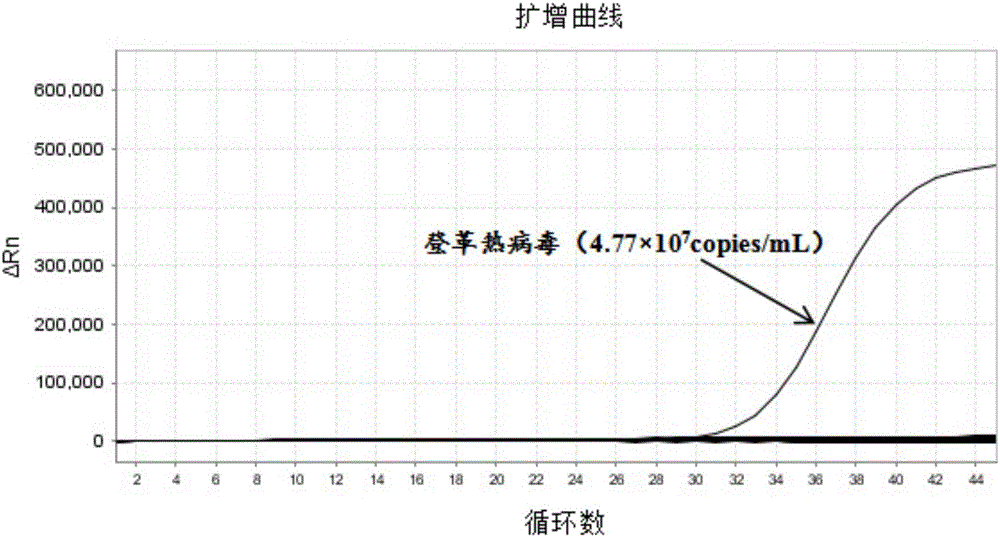
[0090] The standard strain nucleic acid sample of Zika virus is diluted to the nucleic acid concentration of 0.001 LD50; Carry out Zika virus, dengue fever virus, chikungunya fever virus triple real-time fluorescent RT-PCR and single multiplex according to the method and steps in Example 2 For conventional reagent amplification detection comparison, each concentration was repeated 50 times. The results are shown in Table 3 and figure 1 shown.
[0091] Table 3 Analysis sensitivity results
[0092] Detection method Total number of samples (sets) Positive samples (parts) The detection rate(%) Triple Real-Time Fluorescent RT-PCR Kit 50 48 96 Conventional Singleplex RT-PCR Kit 50 44 88
[0093] from Table 3 and figure 1 From the results shown, it can be seen that the analytical ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com