Radix isatidis protein identification method
A technology of protein identification and Radix Radix is applied in the field of analytical chemistry and bioinformatics to achieve the effects of easy mastery, good repeatability and easy operation
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0040] Example 1: Mass spectrometry identification of Radix isatidis protein
[0041] 1. Experimental materials
[0042] 1.1 Instrument
[0043]Thermo Scientific EASY-nLC 1000 nanoliter liquid chromatography (Thermo Company, USA);
[0044] Millipore water purifier ultrapure water system (Millipore, Bedford, MA);
[0045] Desktop high-speed refrigerated centrifuge (Allegra X-30R type, American Beckman Coulter Company);
[0046] SDS-PAGE electrophoresis instrument (USA BIO-RAD company);
[0047] Thermo scientific EASY C18 column (2cm×100μm, 5μm);
[0048] Thermo scientific EASY C18 column (75μm×100mm, 3μm);
[0049] Fully automatic 680 microplate reader (BIO-RAD, USA);
[0050] Q-Exactive mass spectrometer (Thermo Finnigan, USA).
[0051] 1.2 Reagents and reagents
[0052] Trichloroacetic acid (TCA) (Sinopharm Chemical Reagent Co., Ltd.);
[0053] Trypsin (Shanghai Solarbio Biotechnology Co., Ltd.);
[0054] Formic acid, acetonitrile (German Merck company);
[0055] S...
Embodiment 2
[0075] Example 2: Bioinformatics analysis of Radix isatidis protein
[0076] 1 Sequence Information Extraction
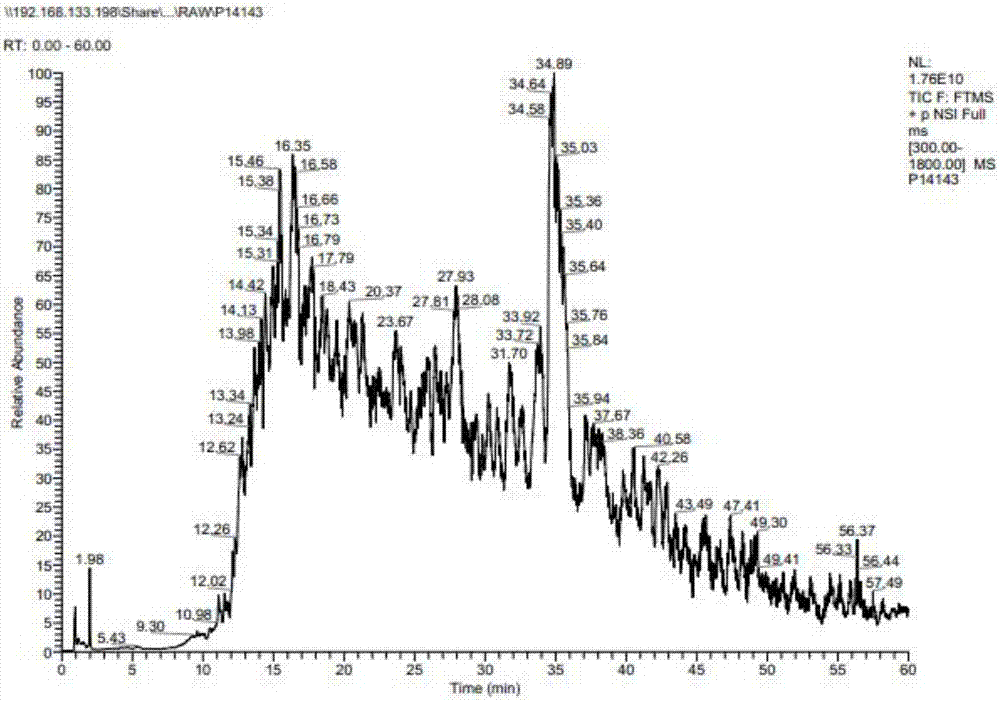
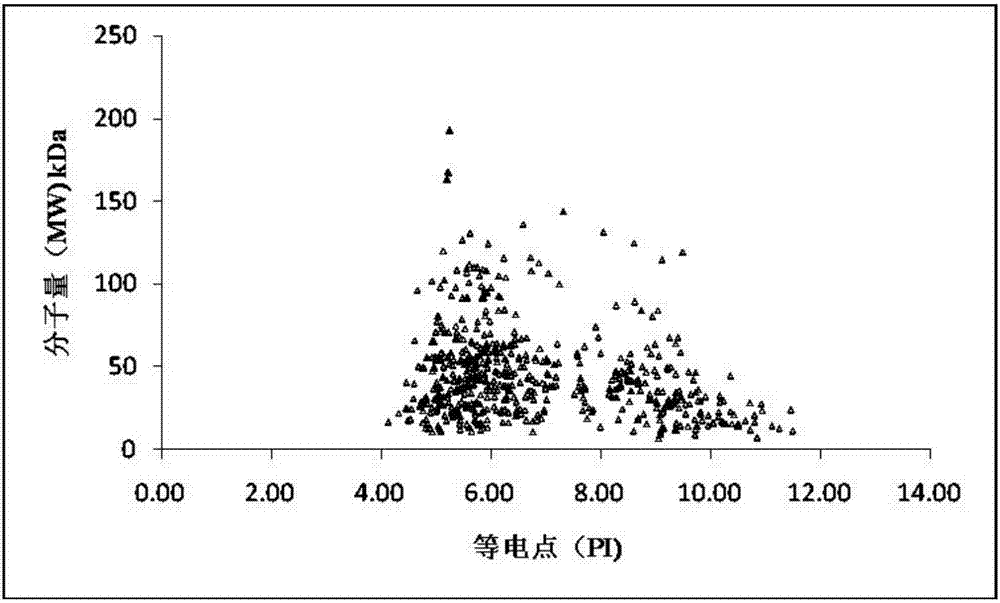
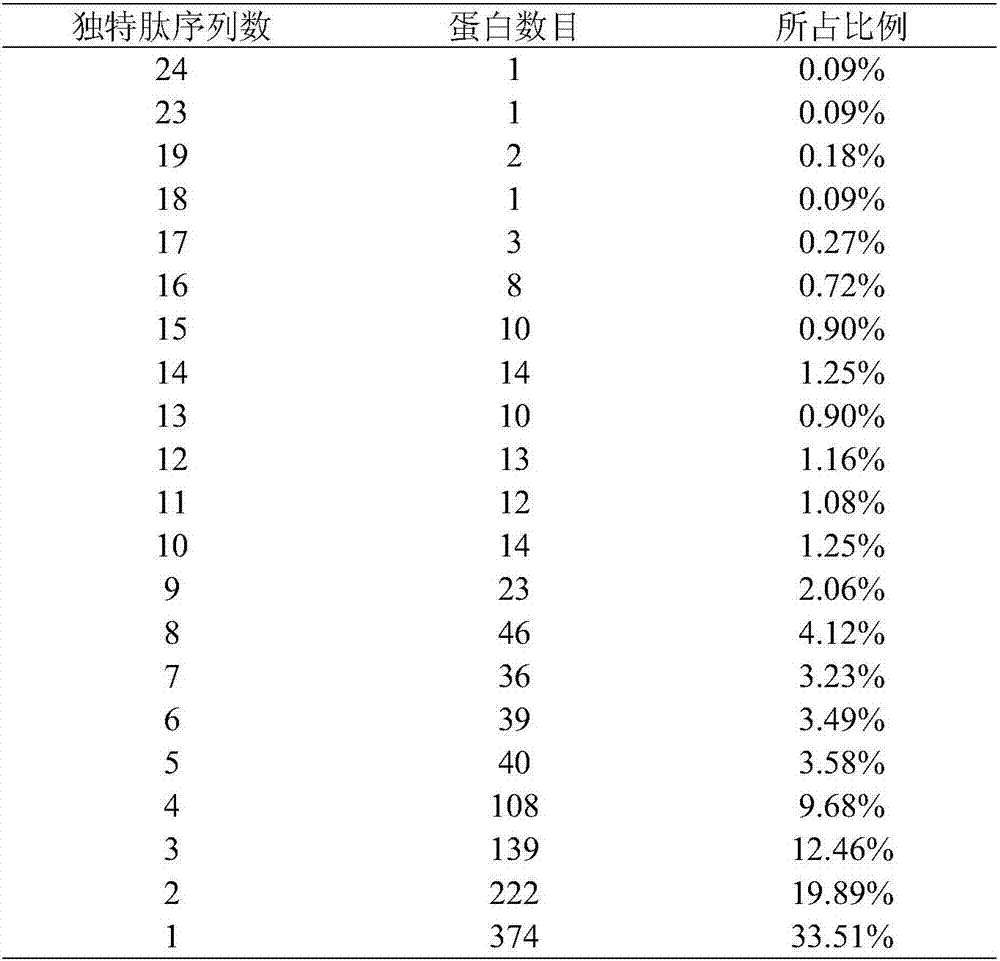
[0077] A total of 1116 proteins were identified by mass spectrometry in the original data (O49485 in the protein identification results was replaced by Q8LGJ6 after the database was updated), and the protein sequence information was extracted from the NCBI database in batches (version number: release 2014_09) and saved in FASTA format (201409121009S0LYNE.fasta ).
[0078] 2 Gene Ontology (Gene Ontology, GO) functional annotation
[0079] 2.1 Sequence Alignment (BLAST)
[0080] The identified proteins were compared with the protein sequences in the NCBI nr database using the localized sequence alignment software NCBI BLAST+ (ncbi-blast-2.2.28+-win32.ext). According to the principle of similarity, the obtained functional information of homologous proteins can be used for functional annotation of target proteins.
[0081] 2.2 GO function entry extraction (Mapping) ...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Molecular weight | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com